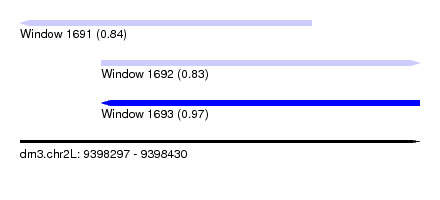
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 9,398,297 – 9,398,430 |
| Length | 133 |
| Max. P | 0.971073 |

| Location | 9,398,297 – 9,398,394 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 75.03 |
| Shannon entropy | 0.46339 |
| G+C content | 0.48437 |
| Mean single sequence MFE | -32.88 |
| Consensus MFE | -17.88 |
| Energy contribution | -18.02 |
| Covariance contribution | 0.15 |
| Combinations/Pair | 1.48 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.836538 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
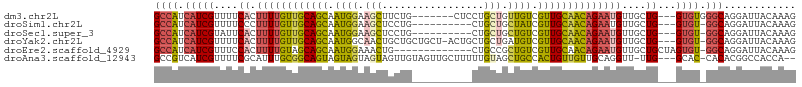
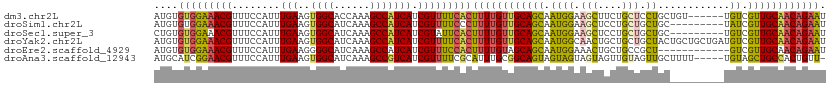
>dm3.chr2L 9398297 97 - 23011544 GCCAUCAUCGUUUUCACUUUUGUUGCAGCAAUGGAAGCUUCUG-------CUCCUGCUGUUGUCGUUGCAACAGAAUGUUGCUG---GUGUGGGCAGGAUUACAAAG (((..((((((...((.((((((((((((...(((.((....)-------)))).((....)).))))))))))))))..)).)---)))..)))............ ( -31.20, z-score = -1.23, R) >droSim1.chr2L 9174396 93 - 22036055 GCCAUCAUCGUUUUCCCUUUUGUUGCAGCAAUGGAAGCUCCUG----------CUGCUGCUAUCGUUGCAACAGAAUGUUGCUG---GUGU-GGCAGGAUUACAAAG .........((..(((.((((((((((((.((((.(((.....----------..))).)))).))))))))))))(((..(..---..).-.))))))..)).... ( -33.50, z-score = -2.48, R) >droSec1.super_3 4852179 93 - 7220098 GCCAUCAUCGUAUUCACUUUUGUUGCAGCAAUGGAAGCUCCUG----------CUGCUGCUGUCGUUGCAACAGAAUGUUGCUG---GUGU-GGCAGGAUUACAAAG ((((.(((((((..((.((((((((((((.((((.(((.....----------..))).)))).)))))))))))))).))).)---))))-)))............ ( -34.70, z-score = -2.39, R) >droYak2.chr2L 12069256 102 - 22324452 GCCAUCAUCGUUUUCACUUUUGUUGCAGCAAUGGCAACUGCUGCUGCU-ACUGCUGCUGAUGUCGUUGCAACAGAAUGUUGCUG---GUGU-GGCAGGAUUACAAAG ((((.((((((...((.((((((((((((...((((.(.((.((....-...)).)).).))))))))))))))))))..)).)---))))-)))............ ( -35.40, z-score = -1.30, R) >droEre2.scaffold_4929 10007572 93 - 26641161 GCCAUCAUCGUUUCCACUUUUGUAGCAGCAAUGGAAACUG-------------CUGCCGCUGUCGUUGCAACAGAAUGUUGCUGCUAGUGU-GGCAGGAUUACAAAG ......((((((((((...((((....)))))))))))..-------------(((((((....((.(((((.....))))).))....))-))))))))....... ( -30.90, z-score = -1.50, R) >droAna3.scaffold_12943 750647 100 - 5039921 GCCGUCAUCGUUUUCGCAUUUGCGGCAGUAGUAGUAGUAGUUGUAGUUGCUUUUUGUAGCUGCCACUGUUGUUGCAGGUU-UUG---GCAC-CACACGGCCACCA-- (((((.((((....)).))..)))))....(((((((((((.((((((((.....)))))))).))))))))))).(((.-..(---((..-......)))))).-- ( -31.60, z-score = -1.32, R) >consensus GCCAUCAUCGUUUUCACUUUUGUUGCAGCAAUGGAAGCUGCUG__________CUGCUGCUGUCGUUGCAACAGAAUGUUGCUG___GUGU_GGCAGGAUUACAAAG (((..(((.......((((((((((((((.((((.(((.................))).)))).)))))))))))).))........)))..)))............ (-17.88 = -18.02 + 0.15)

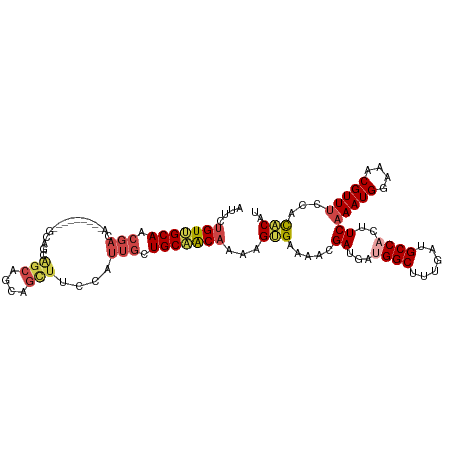
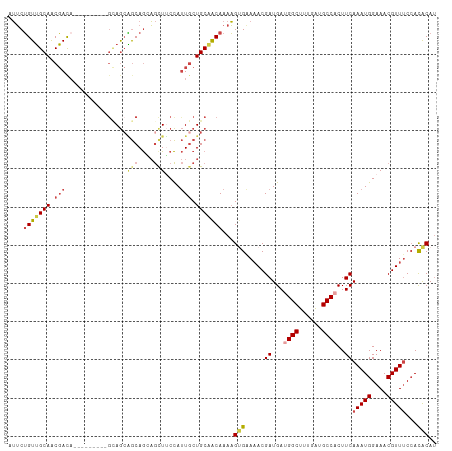
| Location | 9,398,324 – 9,398,430 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 82.48 |
| Shannon entropy | 0.32922 |
| G+C content | 0.44936 |
| Mean single sequence MFE | -30.98 |
| Consensus MFE | -18.02 |
| Energy contribution | -19.22 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.831069 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 9398324 106 + 23011544 AUUCUGUUGCAACGACA------ACAGCAGGAGCAGAAGCUUCCAUUGCUGCAACAAAAGUGAAAACGAUGAUGGCUUUGGUGCCACUUCAAAUGGAAACGUUUCCACACAU ....((((((...(...------.)(((((((((....)))))...))))))))))...(((.((((((.(.((((......))))).))....(....))))).))).... ( -29.40, z-score = -1.35, R) >droSim1.chr2L 9174422 103 + 22036055 AUUCUGUUGCAACGAUA---------GCAGCAGCAGGAGCUUCCAUUGCUGCAACAAAAGGGAAAACGAUGAUGGCUUUGAUGCCACUUCAAAUGGAAACGUUUCCACACAU ....(((((((.((((.---------(.(((.......))).).)))).)))))))....(((((..((.(.((((......))))).))...((....)))))))...... ( -28.60, z-score = -1.41, R) >droSec1.super_3 4852205 103 + 7220098 AUUCUGUUGCAACGACA---------GCAGCAGCAGGAGCUUCCAUUGCUGCAACAAAAGUGAAUACGAUGAUGGCUUUGAUGCCACUUCAAAUGGAAACGUUUCCACACAG ...((((((...)))))---------)..((((((((.....))..)))))).......(((.....((.(.((((......))))).))(((((....)))))...))).. ( -30.00, z-score = -1.75, R) >droYak2.chr2L 12069282 112 + 22324452 AUUCUGUUGCAACGACAUCAGCAGCAGUAGCAGCAGCAGUUGCCAUUGCUGCAACAAAAGUGAAAACGAUGAUGGCUUUGAUGCCACUUCAAAUGGAAACGUUUCCACACAU ....(((((...)))))...((((((((.(((((....))))).)))))))).......(((.((((((.(.((((......))))).))....(....))))).))).... ( -37.00, z-score = -2.58, R) >droEre2.scaffold_4929 10007601 100 + 26641161 AUUCUGUUGCAACGAC------------AGCGGCAGCAGUUUCCAUUGCUGCUACAAAAGUGGAAACGAUGAUGGCUUUGAUGCCCCUUCAAAUGGAAACGUUUCCACACAU ...((((((...))))------------)).(((((((((....)))))))))......((((((((..(((.(((......)))...)))...(....))))))))).... ( -36.60, z-score = -4.07, R) >droAna3.scaffold_12943 750677 106 + 5039921 -AACAGUGGCAGCUACA-----AAAAGCAACUACAACUACUACUACUACUGCCGCAAAUGCGAAAACGAUGACGGCUUUGAUGCCACUUCAAAUGGAAACGUUCCGAUGCAU -...((((((((((...-----...)))......................((((((....((....)).)).)))).....)))))))((.((((....))))..))..... ( -24.30, z-score = -2.15, R) >consensus AUUCUGUUGCAACGACA_________GCAGCAGCAGCAGCUUCCAUUGCUGCAACAAAAGUGAAAACGAUGAUGGCUUUGAUGCCACUUCAAAUGGAAACGUUUCCACACAU ....(((((((.(((...........((....))...........))).)))))))...(((.....((...((((......))))..))(((((....)))))...))).. (-18.02 = -19.22 + 1.20)
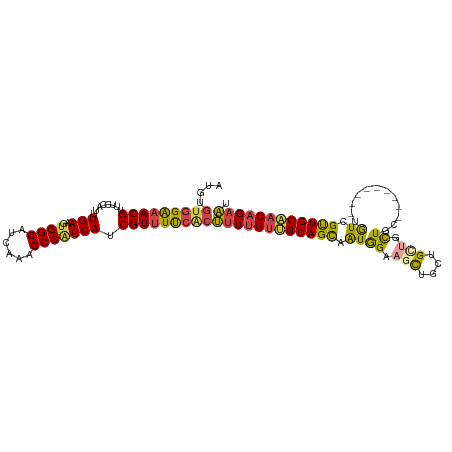
| Location | 9,398,324 – 9,398,430 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 82.48 |
| Shannon entropy | 0.32922 |
| G+C content | 0.44936 |
| Mean single sequence MFE | -34.02 |
| Consensus MFE | -24.78 |
| Energy contribution | -25.20 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.84 |
| SVM RNA-class probability | 0.971073 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
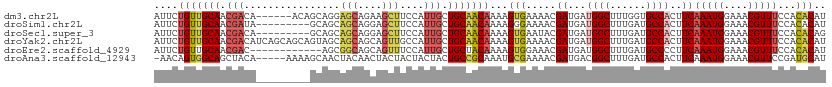
>dm3.chr2L 9398324 106 - 23011544 AUGUGUGGAAACGUUUCCAUUUGAAGUGGCACCAAAGCCAUCAUCGUUUUCACUUUUGUUGCAGCAAUGGAAGCUUCUGCUCCUGCUGU------UGUCGUUGCAACAGAAU ....(((((((((.(((.....)))(((((......)))))...)))))))))((((((((((((...(((.((....))))).((...------.)).)))))))))))). ( -36.00, z-score = -2.89, R) >droSim1.chr2L 9174422 103 - 22036055 AUGUGUGGAAACGUUUCCAUUUGAAGUGGCAUCAAAGCCAUCAUCGUUUUCCCUUUUGUUGCAGCAAUGGAAGCUCCUGCUGCUGC---------UAUCGUUGCAACAGAAU ....((((((....))))))..((.(((((......)))))..))........((((((((((((.((((.(((.......))).)---------))).)))))))))))). ( -33.90, z-score = -2.77, R) >droSec1.super_3 4852205 103 - 7220098 CUGUGUGGAAACGUUUCCAUUUGAAGUGGCAUCAAAGCCAUCAUCGUAUUCACUUUUGUUGCAGCAAUGGAAGCUCCUGCUGCUGC---------UGUCGUUGCAACAGAAU ....(((((.(((.(((.....)))(((((......)))))...))).)))))((((((((((((.((((.(((.......))).)---------))).)))))))))))). ( -34.00, z-score = -2.27, R) >droYak2.chr2L 12069282 112 - 22324452 AUGUGUGGAAACGUUUCCAUUUGAAGUGGCAUCAAAGCCAUCAUCGUUUUCACUUUUGUUGCAGCAAUGGCAACUGCUGCUGCUACUGCUGCUGAUGUCGUUGCAACAGAAU ....(((((((((.(((.....)))(((((......)))))...)))))))))((((((((((((...((((.(.((.((.......)).)).).)))))))))))))))). ( -38.80, z-score = -2.37, R) >droEre2.scaffold_4929 10007601 100 - 26641161 AUGUGUGGAAACGUUUCCAUUUGAAGGGGCAUCAAAGCCAUCAUCGUUUCCACUUUUGUAGCAGCAAUGGAAACUGCUGCCGCU------------GUCGUUGCAACAGAAU ....(((((((((...((.......))(((......))).....)))))))))((((((.((((((..(....))))))).((.------------......)).)))))). ( -33.80, z-score = -2.45, R) >droAna3.scaffold_12943 750677 106 - 5039921 AUGCAUCGGAACGUUUCCAUUUGAAGUGGCAUCAAAGCCGUCAUCGUUUUCGCAUUUGCGGCAGUAGUAGUAGUAGUUGUAGUUGCUUUU-----UGUAGCUGCCACUGUU- .(((...(((.....)))....((.(((((......)))).).)).....(((....))))))..........((((.((((((((....-----.)))))))).))))..- ( -27.60, z-score = -0.84, R) >consensus AUGUGUGGAAACGUUUCCAUUUGAAGUGGCAUCAAAGCCAUCAUCGUUUUCACUUUUGUUGCAGCAAUGGAAGCUGCUGCUGCUGC_________UGUCGUUGCAACAGAAU ....(((((((((........(((..((((......))))))).)))))))))((((((((((((.(((.........((....)).........))).)))))))))))). (-24.78 = -25.20 + 0.42)
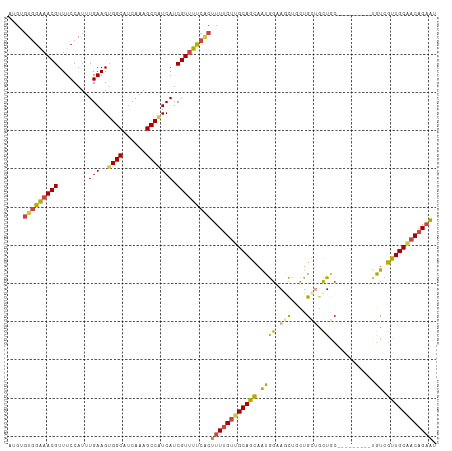
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:28:12 2011