| Sequence ID | dm3.chr2L |
|---|---|
| Location | 842,067 – 842,159 |
| Length | 92 |
| Max. P | 0.860637 |

| Location | 842,067 – 842,159 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 73.86 |
| Shannon entropy | 0.43496 |
| G+C content | 0.58498 |
| Mean single sequence MFE | -34.60 |
| Consensus MFE | -19.72 |
| Energy contribution | -19.76 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.658006 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 842067 92 + 23011544 --------------UGCCCCAAGCCCCAAGACAUGACUCGUUGCUUGGCAACCGAAAGCGGUGUGGUGAUGAUCU--CGGGGAGAUGCAGAAGAAUGACUCUGGGGCC --------------.(((((...((((.((((((.(((((..(((((....)...))))..)).))).))).)))--.))))(((..((......))..)))))))). ( -36.30, z-score = -2.40, R) >droEre2.scaffold_4929 898847 82 + 26641161 ---------------------UGCCCCAAGACAUGACUCGUUGCUUGGCCACCGAAAGGG-----GAGGAGAGAUCUCGCAGAGCUGGAGAGGAAUGGCUCCGGGGCC ---------------------.(((((.((.(((..(((...((((.((..((....)).-----...(((....))))).))))....)))..))).))..))))). ( -34.00, z-score = -1.68, R) >droYak2.chr2L 825677 87 + 22324452 ---------------------UGCCCCAAGACAUGACUCGUUGCUUGGCAACCGUAGAAGAGUGAGAGGAGAGAUCUCGACGAGAUGGAGAAGAAUGACUCUGGGGCC ---------------------.(((((.........((((((((((.((....))....))))((((.......))))))))))..((((........))))))))). ( -27.60, z-score = -1.94, R) >droSec1.super_14 812680 108 + 2068291 UGCCCCAAGCUCCAAGCCCCAAGCCCCAAGACAUGACUCGUUGCUUGGCAACCGAGAGCGGUGUGGAGAUGAUCUCUCGGGGAGAUGCAGAAGAAUGACUCUGGGGCC .((((((..(((((....((((((..(.((......)).)..))))))..((((....)))).))))).(((((((.....))))).))............)))))). ( -36.20, z-score = -0.13, R) >droSim1.chr2L 840931 101 + 22036055 -------UGCCCCAAGCUCCAAGCCCCAAGACAUGACUCGUUGCUUGGCAACCGAGAGCGGUGUGGAGAUGAUCUCUCGGGGAGAUGCAGCAGAAUGACUCUGGGGCC -------.(((((..(((.((..((((....(((.((.((((.(((((...))))))))))))))((((.....))))))))...)).)))(((.....)))))))). ( -38.90, z-score = -1.24, R) >consensus _______________GC_CCAAGCCCCAAGACAUGACUCGUUGCUUGGCAACCGAAAGCGGUGUGGAGAUGAUCUCUCGGGGAGAUGCAGAAGAAUGACUCUGGGGCC ......................(((((.........((((((((...))))).............((((.....))))...))).......(((.....)))))))). (-19.72 = -19.76 + 0.04)

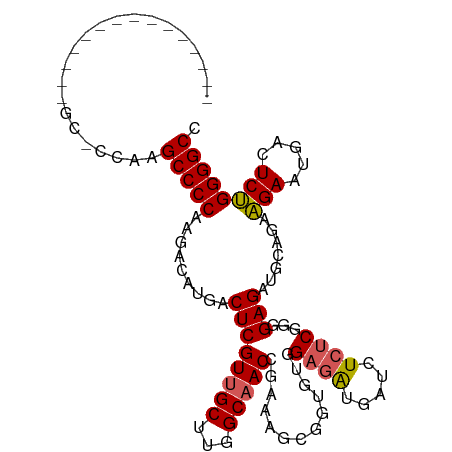
| Location | 842,067 – 842,159 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 73.86 |
| Shannon entropy | 0.43496 |
| G+C content | 0.58498 |
| Mean single sequence MFE | -34.45 |
| Consensus MFE | -22.58 |
| Energy contribution | -23.70 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.860637 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 842067 92 - 23011544 GGCCCCAGAGUCAUUCUUCUGCAUCUCCCCG--AGAUCAUCACCACACCGCUUUCGGUUGCCAAGCAACGAGUCAUGUCUUGGGGCUUGGGGCA-------------- .(((((((((......))))......(((((--(((.(((.(((....((....))(((((...)))))).)).)))))))))))...))))).-------------- ( -32.70, z-score = -1.64, R) >droEre2.scaffold_4929 898847 82 - 26641161 GGCCCCGGAGCCAUUCCUCUCCAGCUCUGCGAGAUCUCUCCUC-----CCCUUUCGGUGGCCAAGCAACGAGUCAUGUCUUGGGGCA--------------------- .(((((((((........)))).((((((((((....)))..(-----(((....)).))....)))..))))........))))).--------------------- ( -27.70, z-score = -0.40, R) >droYak2.chr2L 825677 87 - 22324452 GGCCCCAGAGUCAUUCUUCUCCAUCUCGUCGAGAUCUCUCCUCUCACUCUUCUACGGUUGCCAAGCAACGAGUCAUGUCUUGGGGCA--------------------- .((((((.((.(((....(((.........((((.......))))...........(((((...))))))))..))).)))))))).--------------------- ( -26.10, z-score = -2.13, R) >droSec1.super_14 812680 108 - 2068291 GGCCCCAGAGUCAUUCUUCUGCAUCUCCCCGAGAGAUCAUCUCCACACCGCUCUCGGUUGCCAAGCAACGAGUCAUGUCUUGGGGCUUGGGGCUUGGAGCUUGGGGCA .(((((((((....(((..((........))..)))....)))......(((((.((((.((((((..((((......))))..)))))))))).))))).)))))). ( -45.00, z-score = -1.62, R) >droSim1.chr2L 840931 101 - 22036055 GGCCCCAGAGUCAUUCUGCUGCAUCUCCCCGAGAGAUCAUCUCCACACCGCUCUCGGUUGCCAAGCAACGAGUCAUGUCUUGGGGCUUGGAGCUUGGGGCA------- .((((((((((......)))((......(((((((...............)))))))...((((((..((((......))))..)))))).))))))))).------- ( -40.76, z-score = -1.56, R) >consensus GGCCCCAGAGUCAUUCUUCUGCAUCUCCCCGAGAGAUCAUCUCCACACCGCUUUCGGUUGCCAAGCAACGAGUCAUGUCUUGGGGCUUGG_GC_______________ .((((((.((.(((..(((..........((((((...............))))))(((((...))))))))..))).))))))))...................... (-22.58 = -23.70 + 1.12)



Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:07:08 2011