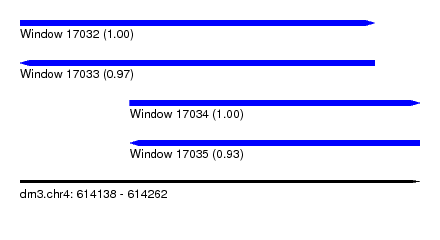
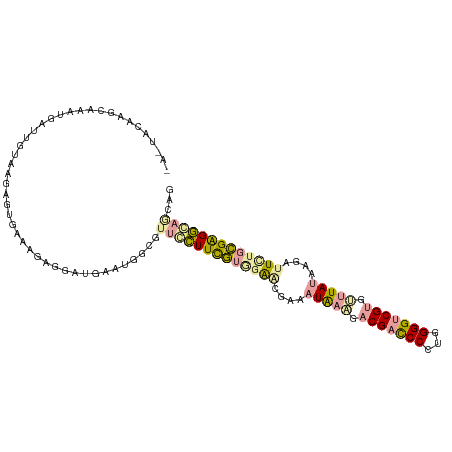
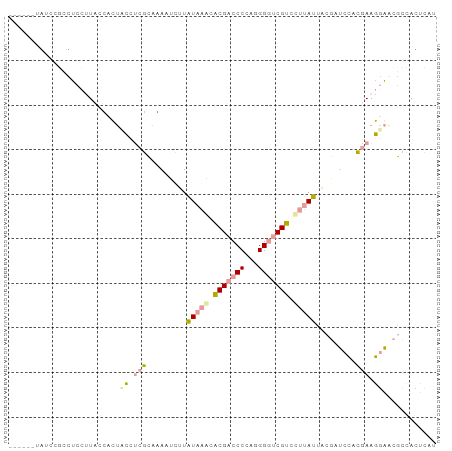
| Sequence ID | dm3.chr4 |
|---|---|
| Location | 614,138 – 614,262 |
| Length | 124 |
| Max. P | 0.999133 |

| Location | 614,138 – 614,248 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 60.30 |
| Shannon entropy | 0.57133 |
| G+C content | 0.49802 |
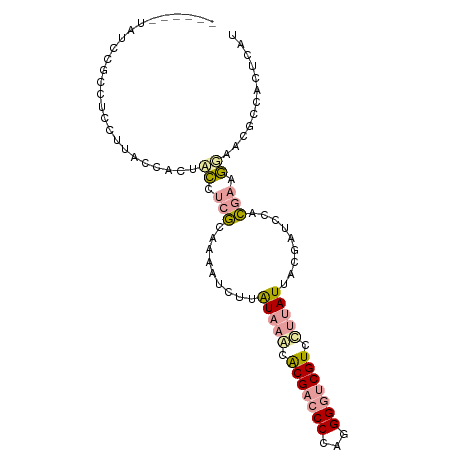
| Mean single sequence MFE | -37.40 |
| Consensus MFE | -18.07 |
| Energy contribution | -19.20 |
| Covariance contribution | 1.13 |
| Combinations/Pair | 1.52 |
| Mean z-score | -3.05 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.66 |
| SVM RNA-class probability | 0.999133 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr4 614138 110 + 1351857 AACUACAAGAAGCUCAUUGGAAGGGCGAAGAUGGAUGACUGGAAACGCUUCGUGGGAGAUCAUGCCGACGACCCAUGGGGGCGCGUCUACAAGAUUUGCCGAGGCCGCAG ..((.(((........)))..)).(((.............(....)(((((..((.(((((.((..((((.(((....)))..))))..)).))))).)))))))))).. ( -30.50, z-score = 0.59, R) >droAna3.scaffold_13089 82066 107 - 978470 ---UACAAGCAUAGGGUUUUGCGUGUGAAAGAUGAUGAGUGGCGUUCUUUCGUAGAUCGAAAUAAACACGAUCCCUGGGGUCGUGUUUAUAAGAUUCUGCGGGAGAAUGG ---((((.(((........))).))))...............((((((..((((((((...(((((((((((((...)))))))))))))..)).))))))..)))))). ( -42.90, z-score = -5.76, R) >droGri2.scaffold_15040 43428 110 - 162499 GAGUACAAGCAAAUGCUUGUAAGAGUGAAAGAGGAUGAAUGGCGUUCCUUUUUGGAGCGCAAUAAGGACGACCCCUGGGGUCGUGCUUAUAAAGUUGUGAGAGGUAGGCG ...(((((((....)))))))...((...............(((((((.....))))))).(((((.(((((((...))))))).))))).................)). ( -38.80, z-score = -3.98, R) >consensus _A_UACAAGCAAAUGAUUGUAAGAGUGAAAGAGGAUGAAUGGCGUUCCUUCGUGGAACGAAAUAAAGACGACCCCUGGGGUCGUGUUUAUAAGAUUCUGCGAGGCAGCAG ............................................(((((((((((((....(((((.(((((((...))))))).)))))....)))))))))))))... (-18.07 = -19.20 + 1.13)

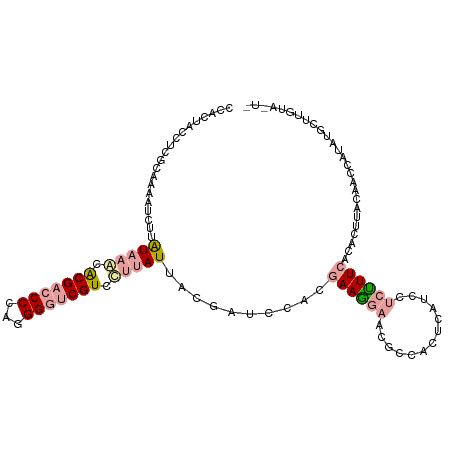
| Location | 614,138 – 614,248 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 60.30 |
| Shannon entropy | 0.57133 |
| G+C content | 0.49802 |
| Mean single sequence MFE | -27.14 |
| Consensus MFE | -8.79 |
| Energy contribution | -10.35 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.80 |
| Structure conservation index | 0.32 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.84 |
| SVM RNA-class probability | 0.970658 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr4 614138 110 - 1351857 CUGCGGCCUCGGCAAAUCUUGUAGACGCGCCCCCAUGGGUCGUCGGCAUGAUCUCCCACGAAGCGUUUCCAGUCAUCCAUCUUCGCCCUUCCAAUGAGCUUCUUGUAGUU ((((((....((...(((.(((.((((.((((....)))))))).))).)))...))..(((((.................................))))))))))).. ( -25.81, z-score = 0.77, R) >droAna3.scaffold_13089 82066 107 + 978470 CCAUUCUCCCGCAGAAUCUUAUAAACACGACCCCAGGGAUCGUGUUUAUUUCGAUCUACGAAAGAACGCCACUCAUCAUCUUUCACACGCAAAACCCUAUGCUUGUA--- ...((((..((.(((.((..((((((((((((....)).))))))))))...))))).))..))))..................(((.(((........))).))).--- ( -25.90, z-score = -4.23, R) >droGri2.scaffold_15040 43428 110 + 162499 CGCCUACCUCUCACAACUUUAUAAGCACGACCCCAGGGGUCGUCCUUAUUGCGCUCCAAAAAGGAACGCCAUUCAUCCUCUUUCACUCUUACAAGCAUUUGCUUGUACUC ....................(((((.(((((((...))))))).))))).(((.(((.....))).)))....................(((((((....)))))))... ( -29.70, z-score = -4.94, R) >consensus CCACUACCUCGCAAAAUCUUAUAAACACGACCCCAGGGGUCGUCCUUAUUACGAUCCACGAAGGAACGCCACUCAUCCUCUUUCACACUUACAACCAUAUGCUUGUA_U_ ....................(((((.(((((((...))))))).)))))..........((((((.............)))))).......................... ( -8.79 = -10.35 + 1.56)

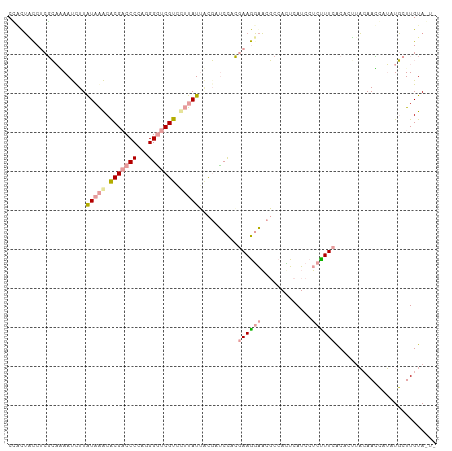
| Location | 614,172 – 614,262 |
|---|---|
| Length | 90 |
| Sequences | 3 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 55.67 |
| Shannon entropy | 0.62163 |
| G+C content | 0.53356 |
| Mean single sequence MFE | -33.43 |
| Consensus MFE | -18.07 |
| Energy contribution | -19.20 |
| Covariance contribution | 1.13 |
| Combinations/Pair | 1.52 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.65 |
| SVM RNA-class probability | 0.999110 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr4 614172 90 + 1351857 AUGACUGGAAACGCUUCGUGGGAGAUCAUGCCGACGACCCAUGGGGGCGCGUCUACAAGAUUUGCCGAGGCCGCAGAAAGUGCACGGAGA------ ......(....).((((((((.(((((.((..((((.(((....)))..))))..)).))))).))......(((.....))))))))).------ ( -29.30, z-score = -0.35, R) >droAna3.scaffold_13089 82097 96 - 978470 AUGAGUGGCGUUCUUUCGUAGAUCGAAAUAAACACGAUCCCUGGGGUCGUGUUUAUAAGAUUCUGCGGGAGAAUGGGAAGGAGGGGUCCAGGGAUA ........((((((..((((((((...(((((((((((((...)))))))))))))..)).))))))..))))))....(((....)))....... ( -39.60, z-score = -4.78, R) >droGri2.scaffold_15040 43462 90 - 162499 AUGAAUGGCGUUCCUUUUUGGAGCGCAAUAAGGACGACCCCUGGGGUCGUGCUUAUAAAGUUGUGAGAGGUAGGCGUAGGGAAGCGGAUG------ .......(((((((.....))))))).(((((.(((((((...))))))).))))).................((........)).....------ ( -31.40, z-score = -2.43, R) >consensus AUGAAUGGCGUUCCUUCGUGGAACGAAAUAAAGACGACCCCUGGGGUCGUGUUUAUAAGAUUCUGCGAGGCAGCAGAAAGGAAGCGGACA______ ..........(((((((((((((....(((((.(((((((...))))))).)))))....)))))))))))))....................... (-18.07 = -19.20 + 1.13)


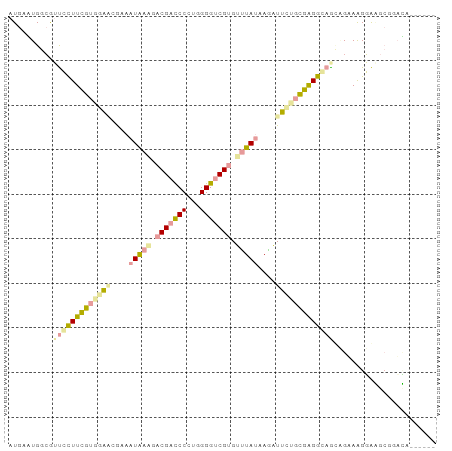
| Location | 614,172 – 614,262 |
|---|---|
| Length | 90 |
| Sequences | 3 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 55.67 |
| Shannon entropy | 0.62163 |
| G+C content | 0.53356 |
| Mean single sequence MFE | -22.77 |
| Consensus MFE | -8.60 |
| Energy contribution | -9.50 |
| Covariance contribution | 0.90 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.928944 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr4 614172 90 - 1351857 ------UCUCCGUGCACUUUCUGCGGCCUCGGCAAAUCUUGUAGACGCGCCCCCAUGGGUCGUCGGCAUGAUCUCCCACGAAGCGUUUCCAGUCAU ------.....(((.(((....(((.(.(((....(((.(((.((((.((((....)))))))).))).)))......))).))))....)))))) ( -23.20, z-score = 0.57, R) >droAna3.scaffold_13089 82097 96 + 978470 UAUCCCUGGACCCCUCCUUCCCAUUCUCCCGCAGAAUCUUAUAAACACGACCCCAGGGAUCGUGUUUAUUUCGAUCUACGAAAGAACGCCACUCAU .......(((....)))......((((..((.(((.((..((((((((((((....)).))))))))))...))))).))..)))).......... ( -23.50, z-score = -3.38, R) >droGri2.scaffold_15040 43462 90 + 162499 ------CAUCCGCUUCCCUACGCCUACCUCUCACAACUUUAUAAGCACGACCCCAGGGGUCGUCCUUAUUGCGCUCCAAAAAGGAACGCCAUUCAU ------.....((........)).................(((((.(((((((...))))))).))))).(((.(((.....))).)))....... ( -21.60, z-score = -3.06, R) >consensus ______UAUCCGCCUCCUUACCACUACCUCGCAAAAUCUUAUAAACACGACCCCAGGGGUCGUCCUUAUUACGAUCCACGAAGGAACGCCACUCAU .........................((.(((.........(((((.(((((((...))))))).))))).........))).))............ ( -8.60 = -9.50 + 0.90)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:01:19 2011