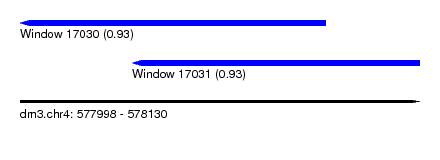
| Sequence ID | dm3.chr4 |
|---|---|
| Location | 577,998 – 578,130 |
| Length | 132 |
| Max. P | 0.934980 |

| Location | 577,998 – 578,099 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.44 |
| Shannon entropy | 0.35749 |
| G+C content | 0.36909 |
| Mean single sequence MFE | -17.00 |
| Consensus MFE | -12.22 |
| Energy contribution | -12.70 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.934980 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
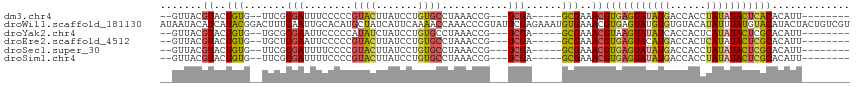
>dm3.chr4 577998 101 - 1351857 CUUAUCCUGUGCCUAAACCG---UCGA-----GCGAAACGUGAGUAUAUGACCACCUAUAUACUCACACAUU----UUCUACAUA-AAUACAAAUACACGCAAACUUAUUUUUG------ .......((((.......((---....-----.))....(((((((((((......))))))))))).....----.........-............))))............------ ( -17.00, z-score = -3.06, R) >droWil1.scaffold_181130 15262474 120 + 16660200 AUCAUUCAAAACCAAACCCGUAUUCGAGAAAUGUGAAACGUGAGUAUGUGUGUACAUAUUUAUGUACAUACUACUGUCGUGCAUGUUAUGUAAAUACAAACAGACAUACAAAUACGUACA ..................((((((.(...........(((..((((.((((((((((....))))))))))))))..)))..(((((.(((........)))))))).).)))))).... ( -25.90, z-score = -0.61, R) >droYak2.chr4 636861 101 - 1374474 UCUAUCCUGUGCCUAAACCG---UCGA-----GCGAAACGUAAGUAUAUCACCACUCAUAUACUCGCACAUU----UUCUACAUA-AAUACAAAUACCCGCAAACUUAUUUUUG------ .......(((((......((---....-----.)).......(((((((........))))))).)))))..----.........-............................------ ( -10.40, z-score = -1.06, R) >droEre2.scaffold_4512 590703 101 + 1286254 CUUAUCCUGUGCCUAAACCG---UCGA-----GCGAAACGUGAGUACAUGACCACUCAUAUACUCGCACAUU----UUCUACAUA-AAUACAAAUACACGCAAACUUAUUUUUG------ .......((((.......((---....-----.))....(((((((.((((....)))).))))))).....----.........-............))))............------ ( -15.50, z-score = -1.82, R) >droSec1.super_30 302431 101 - 664411 CUUAUCCUGUGCCUAAACCG---UCGA-----GCGAAACGUGAGUAUAUGACCACCUAUAUACUCGCACAUU----UUCUACAUA-AAUACAAAUACACGCAAACUUAUUUUUG------ .......((((.......((---....-----.))....(((((((((((......))))))))))).....----.........-............))))............------ ( -16.60, z-score = -2.43, R) >droSim1.chr4 459563 101 - 949497 CUUAUCCUGUGCCUAAACCG---UCGA-----GCGAAACGUGAGUAUAUGACCACCUAUAUACUCGCACAUU----UUCUACAUA-AAUACAAAUACACGCAAACUUAUUUUUG------ .......((((.......((---....-----.))....(((((((((((......))))))))))).....----.........-............))))............------ ( -16.60, z-score = -2.43, R) >consensus CUUAUCCUGUGCCUAAACCG___UCGA_____GCGAAACGUGAGUAUAUGACCACCUAUAUACUCGCACAUU____UUCUACAUA_AAUACAAAUACACGCAAACUUAUUUUUG______ .........(((...........(((.......)))...(((((((((((......)))))))))))................................))).................. (-12.22 = -12.70 + 0.48)

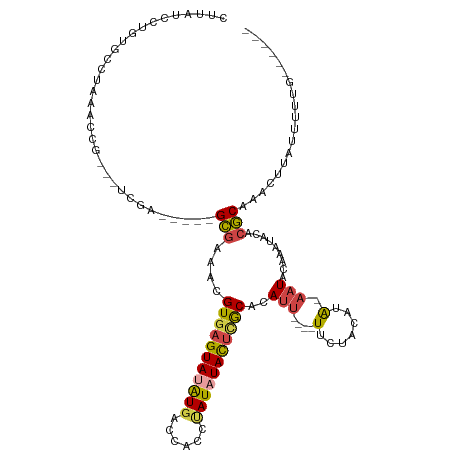
| Location | 578,035 – 578,130 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 75.87 |
| Shannon entropy | 0.41063 |
| G+C content | 0.44744 |
| Mean single sequence MFE | -22.97 |
| Consensus MFE | -12.95 |
| Energy contribution | -13.23 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.934171 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr4 578035 95 - 1351857 --GUUACGUACUGUG--UUCGCGAUUUCCCCCGUACUUAUCCUGUGCCUAAACCG---UCGA-----GCGAAACGUGAGUAUAUGACCACCUAUAUACUCACACAUU-------- --.....((....((--((((((.(((.....((((.......))))..))).))---.)))-----)))..))(((((((((((......))))))))))).....-------- ( -24.20, z-score = -3.22, R) >droWil1.scaffold_181130 15262514 115 + 16660200 AUAAUACAUCAUACGGACUUUCAAUUGCACAUGCUAUCAUUCAAAACCAAACCCGUAUUCGAGAAAUGUGAAACGUGAGUAUGUGUGUACAUAUUUAUGUACAUACUACUGUCGU ...............(((..(((....(((((....((......................))...))))).....)))(((.((((((((((....))))))))))))).))).. ( -22.35, z-score = -0.16, R) >droYak2.chr4 636898 95 - 1374474 --GUUACGUACUGUG--UGCGCGAAUUCCCCCAUAUCUAUCCUGUGCCUAAACCG---UCGA-----GCGAAACGUAAGUAUAUCACCACUCAUAUACUCGCACAUU-------- --(((.((.((.((.--.(((((...................)))))....)).)---))))-----)).....(..(((((((........)))))))..).....-------- ( -14.91, z-score = -0.15, R) >droEre2.scaffold_4512 590740 95 + 1286254 --GUUACGUACUGUG--UGCUCGAAUUCCCCCGUACUUAUCCUGUGCCUAAACCG---UCGA-----GCGAAACGUGAGUACAUGACCACUCAUAUACUCGCACAUU-------- --..........(((--(((((((........((((.......))))........---))))-----)).....(((((((.((((....)))).))))))))))).-------- ( -27.39, z-score = -3.78, R) >droSec1.super_30 302468 95 - 664411 --GUUACGUACUGUG--UUCGCGAUUUUCCCCGUACUUAUCCUGUGCCUAAACCG---UCGA-----GCGAAACGUGAGUAUAUGACCACCUAUAUACUCGCACAUU-------- --.....((....((--((((((.(((.....((((.......))))..))).))---.)))-----)))..))(((((((((((......))))))))))).....-------- ( -24.50, z-score = -2.86, R) >droSim1.chr4 459600 95 - 949497 --GUUACGUACUGUG--UUCGCGAUUUUCCCCGUACUUAUCCUGUGCCUAAACCG---UCGA-----GCGAAACGUGAGUAUAUGACCACCUAUAUACUCGCACAUU-------- --.....((....((--((((((.(((.....((((.......))))..))).))---.)))-----)))..))(((((((((((......))))))))))).....-------- ( -24.50, z-score = -2.86, R) >consensus __GUUACGUACUGUG__UUCGCGAUUUCCCCCGUACUUAUCCUGUGCCUAAACCG___UCGA_____GCGAAACGUGAGUAUAUGACCACCUAUAUACUCGCACAUU________ ..(((.(((.......................((((.......))))......((....))......))).)))(((((((((((......)))))))))))............. (-12.95 = -13.23 + 0.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:01:16 2011