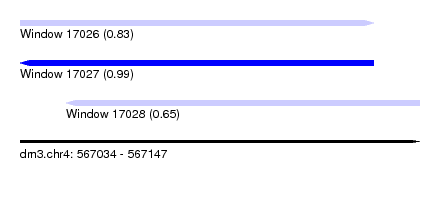
| Sequence ID | dm3.chr4 |
|---|---|
| Location | 567,034 – 567,147 |
| Length | 113 |
| Max. P | 0.989037 |

| Location | 567,034 – 567,134 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 65.92 |
| Shannon entropy | 0.56032 |
| G+C content | 0.55476 |
| Mean single sequence MFE | -30.74 |
| Consensus MFE | -17.90 |
| Energy contribution | -18.50 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.37 |
| Mean z-score | -1.04 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.827384 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr4 567034 100 + 1351857 GGAGGGGAUGUUGAUGCUGGUGCUGCAAGCUCAAGAUACGACUUUCCACAGCAGCCGCAGCUGCCGCUACUGUCGCUAAUGGUAUAUUGCUGCAAUCUUG ((((((.((.((((.(((.(.....).))))))).))....)))))).(((((((.((....)).))((((((.....))))))...)))))........ ( -27.00, z-score = 1.60, R) >droVir3.scaffold_13052 895611 76 - 2019633 ------------------------GCAGGCCGAGUAAGCGGCUUUCAACAGCGGCGGCAGCUGCAGCUGCCGUUGCCGAGGAUAUUGCACUUGAUUCGUG ------------------------((((((((......))))).((..(.(((((((((((....))))))))))).)..))...)))............ ( -33.00, z-score = -1.82, R) >droWil1.scaffold_181130 15251796 83 - 16660200 -----------------UGCUGUUGUAGUCCCAGGAGUCGACUUUCGACAGCAGCUGCAGCGGCCGCAGCUGUCGCCGAUGAUAUGCUAUUCGAUUCAUG -----------------.(((((((.((((.........))))..)))))))(((..((.((((.((....)).)))).))....)))............ ( -25.90, z-score = -0.12, R) >dp4.Unknown_group_78 74768 83 - 77267 -----------------UGUUGGUGUAGACCCAAUAAUCGACUUUCAACAGCGGCAGCAGCAGCUGCUGCCGUUGCUGGUGGUAUGGGGGUGGAAUCAUG -----------------((((((.(....))))))).((.(((..((.((((((((((((...))))))))))))(....)...))..))).))...... ( -33.90, z-score = -2.43, R) >droPer1.super_42 291161 83 - 623919 -----------------UGUUGGUGUAGACCCAAUAAUCGACUUUCAACAGCGGCAGCAGCAGCUGCUGCCGUUGCUGGUGGUAUGGGGGUGGAAUCAUG -----------------((((((.(....))))))).((.(((..((.((((((((((((...))))))))))))(....)...))..))).))...... ( -33.90, z-score = -2.43, R) >consensus _________________UGUUGGUGUAGACCCAAUAAUCGACUUUCAACAGCGGCAGCAGCAGCCGCUGCCGUUGCUGAUGGUAUGGGGCUGGAAUCAUG ..............................(((.......(((.(((.((((((((((.......)))))))))).))).))).......)))....... (-17.90 = -18.50 + 0.60)

| Location | 567,034 – 567,134 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 65.92 |
| Shannon entropy | 0.56032 |
| G+C content | 0.55476 |
| Mean single sequence MFE | -27.60 |
| Consensus MFE | -19.06 |
| Energy contribution | -18.22 |
| Covariance contribution | -0.84 |
| Combinations/Pair | 1.57 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.35 |
| SVM RNA-class probability | 0.989037 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr4 567034 100 - 1351857 CAAGAUUGCAGCAAUAUACCAUUAGCGACAGUAGCGGCAGCUGCGGCUGCUGUGGAAAGUCGUAUCUUGAGCUUGCAGCACCAGCAUCAACAUCCCCUCC ...(((.((..(((.((((.(((..(.((((((((.((....)).)))))))).)..))).)))).))).((.....))....)))))............ ( -30.60, z-score = -0.80, R) >droVir3.scaffold_13052 895611 76 + 2019633 CACGAAUCAAGUGCAAUAUCCUCGGCAACGGCAGCUGCAGCUGCCGCCGCUGUUGAAAGCCGCUUACUCGGCCUGC------------------------ ............(((...((..((((..(((((((....)))))))..))))..))..((((......)))).)))------------------------ ( -27.50, z-score = -1.16, R) >droWil1.scaffold_181130 15251796 83 + 16660200 CAUGAAUCGAAUAGCAUAUCAUCGGCGACAGCUGCGGCCGCUGCAGCUGCUGUCGAAAGUCGACUCCUGGGACUACAACAGCA----------------- .............((......((((((.(((((((((...))))))))).)))))).((((..(....).))))......)).----------------- ( -28.10, z-score = -1.37, R) >dp4.Unknown_group_78 74768 83 + 77267 CAUGAUUCCACCCCCAUACCACCAGCAACGGCAGCAGCUGCUGCUGCCGCUGUUGAAAGUCGAUUAUUGGGUCUACACCAACA----------------- ......................(((((.(((((((((...))))))))).)))))...........((((.......))))..----------------- ( -25.90, z-score = -2.25, R) >droPer1.super_42 291161 83 + 623919 CAUGAUUCCACCCCCAUACCACCAGCAACGGCAGCAGCUGCUGCUGCCGCUGUUGAAAGUCGAUUAUUGGGUCUACACCAACA----------------- ......................(((((.(((((((((...))))))))).)))))...........((((.......))))..----------------- ( -25.90, z-score = -2.25, R) >consensus CAUGAUUCCACCACCAUACCAUCAGCAACGGCAGCAGCAGCUGCUGCCGCUGUUGAAAGUCGAUUAUUGGGCCUACACCAACA_________________ ......................(((((.(((((((((...))))))))).)))))............................................. (-19.06 = -18.22 + -0.84)
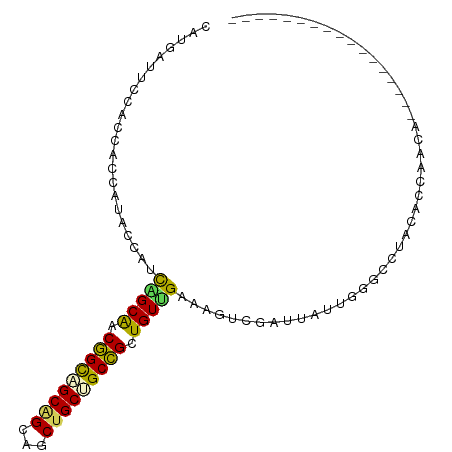
| Location | 567,047 – 567,147 |
|---|---|
| Length | 100 |
| Sequences | 8 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 67.63 |
| Shannon entropy | 0.61012 |
| G+C content | 0.52694 |
| Mean single sequence MFE | -29.69 |
| Consensus MFE | -14.61 |
| Energy contribution | -14.54 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.36 |
| Mean z-score | -0.93 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.645754 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
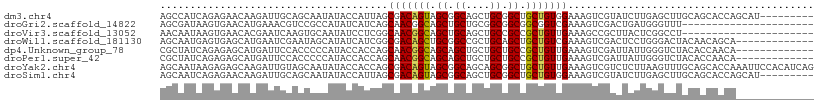
>dm3.chr4 567047 100 - 1351857 AGCCAUCAGAGAACAAGAUUGCAGCAAUAUACCAUUAGCGACAGUAGCGGCAGCUGCGGCUGCUGUGGAAAGUCGUAUCUUGAGCUUGCAGCACCAGCAU--------- ...................((((.(((.((((.(((..(.((((((((.((....)).)))))))).)..))).)))).)))....))))((....))..--------- ( -29.90, z-score = 0.02, R) >droGri2.scaffold_14822 986873 87 - 1097346 AGCGAUAAGUGAACAUGAAACGUCCGCCAUAUCAUCAGCAACGGCAGCUGCUGCGGCGGCGGCGGUCGAAAGUCGACUGAUGGGUUU---------------------- ((((((..(....)......((.(((((.........((....)).(((((....)))))))))).))...)))).)).........---------------------- ( -27.70, z-score = 0.02, R) >droVir3.scaffold_13052 895613 87 + 2019633 AACAAUAAGUGAACACGAAUCAAGUGCAAUAUCCUCGGCAACGGCAGCUGCAGCUGCCGCCGCUGUUGAAAGCCGCUUACUCGGCCU---------------------- .....((((((..(((.......))).....((..((((..(((((((....)))))))..))))..))....))))))........---------------------- ( -27.80, z-score = -1.24, R) >droWil1.scaffold_181130 15251796 96 + 16660200 AGCAAUGAGUGAGCAUGAAUCGAAUAGCAUAUCAUCGGCGACAGCUGCGGCCGCUGCAGCUGCUGUCGAAAGUCGACUCCUGGGACUACAACAGCA------------- .((...((((..((.......((........)).((((((.(((((((((...))))))))).))))))..))..)))).((......))...)).------------- ( -29.50, z-score = 0.22, R) >dp4.Unknown_group_78 74768 96 + 77267 CGCUAUCAGAGAGCAUGAUUCCACCCCCAUACCACCAGCAACGGCAGCAGCUGCUGCUGCCGCUGUUGAAAGUCGAUUAUUGGGUCUACACCAACA------------- .(((.......))).....................(((((.(((((((((...))))))))).)))))...........((((.......))))..------------- ( -28.10, z-score = -1.40, R) >droPer1.super_42 291161 96 + 623919 CGCUAUCAGAGAGCAUGAUUCCACCCCCAUACCACCAGCAACGGCAGCAGCUGCUGCUGCCGCUGUUGAAAGUCGAUUAUUGGGUCUACACCAACA------------- .(((.......))).....................(((((.(((((((((...))))))))).)))))...........((((.......))))..------------- ( -28.10, z-score = -1.40, R) >droYak2.chr4 624947 109 - 1374474 AGCAAUAAGAGAGCAAGAUUGUAGCAAUAUACCACCAGCGACAGUAGCGGCAGCAGCGGCUGCUGUUGAAAGUCGUCUCUUAAGUUUGCAGCACCAAAUUCCACAUCAG .(((((((((((....(((((((....)))........((((((((((.((....)).))))))))))..)))).)))))))...)))).................... ( -32.90, z-score = -2.47, R) >droSim1.chr4 449425 100 - 949497 AGCAAUCAGAGAACAAGAUUGCAGCAAUAUACCAUUAGCGACAGUAGCGGCAGCUGCGGCUGCUGUGGAAAGUCGUAUCUUGAGCUUGCAGCACCAGCAU--------- .((((((.........))))))..(((.((((.(((..(.((((((((.((....)).)))))))).)..))).)))).))).(((.(.....).)))..--------- ( -33.50, z-score = -1.23, R) >consensus AGCAAUCAGAGAACAUGAUUCCAGCACCAUACCAUCAGCAACAGCAGCGGCAGCUGCGGCUGCUGUUGAAAGUCGACUCUUGGGUCUACAGCACCA_____________ ......................................(((((((.((.((....)).)).)))))))......................................... (-14.61 = -14.54 + -0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:01:14 2011