
| Sequence ID | dm3.chr4 |
|---|---|
| Location | 269,187 – 269,296 |
| Length | 109 |
| Max. P | 0.999490 |

| Location | 269,187 – 269,296 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 49.78 |
| Shannon entropy | 0.79651 |
| G+C content | 0.52196 |
| Mean single sequence MFE | -39.08 |
| Consensus MFE | -19.12 |
| Energy contribution | -19.88 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.59 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.94 |
| SVM RNA-class probability | 0.999490 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
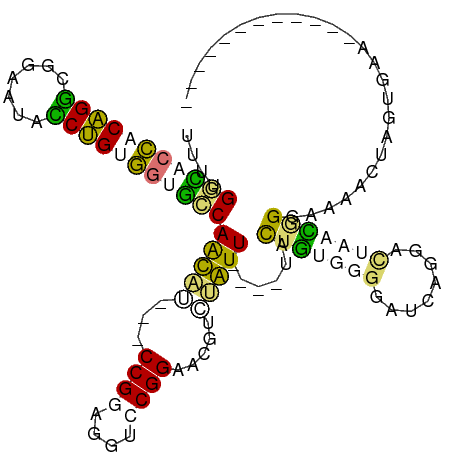
>dm3.chr4 269187 109 + 1351857 UUUUGGUCCUGCAGAUUGUAUUUCUGAAGUGCCAACUUGUGCCGUUAGUCCGGACUCAAUGUU---CCGUUGUGGUGGUAA--ACUAAUCGGGA--CAGGUGGAAUCACUGGUUAA ....((..((.((((.......)))).))..))((((.((((((......)))..(((.((((---(((...((((.....--))))..)))))--))..)))...))).)))).. ( -31.90, z-score = -1.08, R) >droAna3.scaffold_13034 840494 116 + 2223656 UUUUGGUGCCACGGUCGGUAAUGCCGCGAAACCAGCCCGCUCCGUUGGUCCGGGCAGUGGGUUACCCUGCUACGGGGGCCAUUACUACUAGGGAAUUCCAUGGAAUCACUUGGAGU ...((((.((((((((((.....)))....))).(((((..((...))..))))).))))....(((((...)))))))))..(((.(((((..((((....))))..)))))))) ( -44.60, z-score = -0.49, R) >dp4.Unknown_singleton_756 2150 97 - 2355 UUUAGUCACCACAGGCCUAGUACCUGUGGUGGCAAUAU---CCGGAGGUCCGGAACGUCUAUU----UACUGUUGGGAUCAGGAUAGACGGGAAAACUAGCGAA------------ ....(((((((((((.......)))))))))))....(---((((....))))).((((((((----..(((.......)))))))))))..............------------ ( -40.10, z-score = -4.06, R) >droPer1.super_175 73391 97 - 83733 UUUGGGCACCACAGGCGUAGUACCUGUGGUGGCAAUAU---CCGGAGGUCCGGAACGUCUAUU----UACUGGUGGGAUCGGGAUGGACGGGACAACUAGUGAG------------ ....(.(((((((((.......))))))))).)....(---((((....)))))........(----(((((((.(..(((.......)))..).)))))))).------------ ( -39.70, z-score = -3.06, R) >consensus UUUUGGCACCACAGGCGGAAUACCUGUGGUGCCAACAU___CCGGAGGUCCGGAACGUCUAUU____UACUGUGGGGAUCAGGACUAACGGGAAAACUAGUGAA____________ ....(((((((((((.......)))))))))))(((((...((((....)))).....)))))......(((((((........)))))))......................... (-19.12 = -19.88 + 0.75)


| Location | 269,187 – 269,296 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 49.78 |
| Shannon entropy | 0.79651 |
| G+C content | 0.52196 |
| Mean single sequence MFE | -30.98 |
| Consensus MFE | -16.86 |
| Energy contribution | -17.55 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.70 |
| SVM RNA-class probability | 0.999190 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
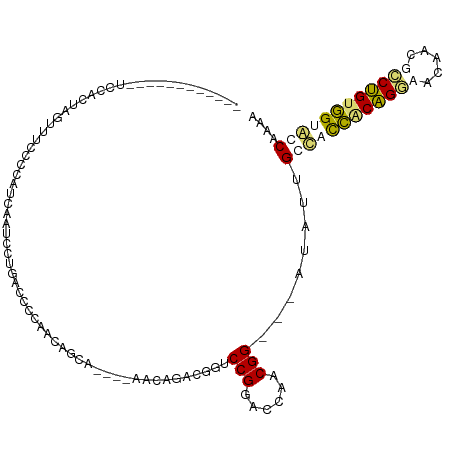
>dm3.chr4 269187 109 - 1351857 UUAACCAGUGAUUCCACCUG--UCCCGAUUAGU--UUACCACCACAACGG---AACAUUGAGUCCGGACUAACGGCACAAGUUGGCACUUCAGAAAUACAAUCUGCAGGACCAAAA .......(((....))).((--(.(((.(((((--(...........(((---(........))))))))))))).)))..((((..((.((((.......)))).))..)))).. ( -23.60, z-score = -0.80, R) >droAna3.scaffold_13034 840494 116 - 2223656 ACUCCAAGUGAUUCCAUGGAAUUCCCUAGUAGUAAUGGCCCCCGUAGCAGGGUAACCCACUGCCCGGACCAACGGAGCGGGCUGGUUUCGCGGCAUUACCGACCGUGGCACCAAAA (((.(.((.(((((....)))))..)).).)))...(((((.(...((((((....)).))))(((......))).).)))))(((.(((((((......).)))))).))).... ( -36.90, z-score = 0.39, R) >dp4.Unknown_singleton_756 2150 97 + 2355 ------------UUCGCUAGUUUUCCCGUCUAUCCUGAUCCCAACAGUA----AAUAGACGUUCCGGACCUCCGG---AUAUUGCCACCACAGGUACUAGGCCUGUGGUGACUAAA ------------...((.........(((((((.(((.......)))..----.))))))).(((((....))))---)....))((((((((((.....))))))))))...... ( -36.00, z-score = -4.64, R) >droPer1.super_175 73391 97 + 83733 ------------CUCACUAGUUGUCCCGUCCAUCCCGAUCCCACCAGUA----AAUAGACGUUCCGGACCUCCGG---AUAUUGCCACCACAGGUACUACGCCUGUGGUGCCCAAA ------------..............((((...................----....)))).(((((....))))---)..(((.((((((((((.....))))))))))..))). ( -27.40, z-score = -2.24, R) >consensus ____________UCCACUAGUUUCCCCAUCAAUCCUGACCCCAACAGCA____AACAGACGGUCCGGACCAACGG___AUAUUGCCACCACAGGAACAACGCCUGUGGUACCAAAA ...............................................................(((......)))........((((((((((((.....)))))))))))).... (-16.86 = -17.55 + 0.69)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:01:03 2011