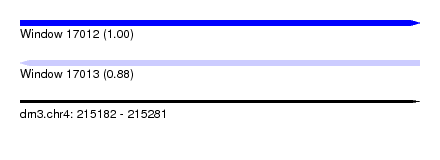
| Sequence ID | dm3.chr4 |
|---|---|
| Location | 215,182 – 215,281 |
| Length | 99 |
| Max. P | 0.999428 |

| Location | 215,182 – 215,281 |
|---|---|
| Length | 99 |
| Sequences | 3 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 57.86 |
| Shannon entropy | 0.54179 |
| G+C content | 0.29348 |
| Mean single sequence MFE | -18.83 |
| Consensus MFE | -9.15 |
| Energy contribution | -10.93 |
| Covariance contribution | 1.78 |
| Combinations/Pair | 1.17 |
| Mean z-score | -3.34 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.88 |
| SVM RNA-class probability | 0.999428 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
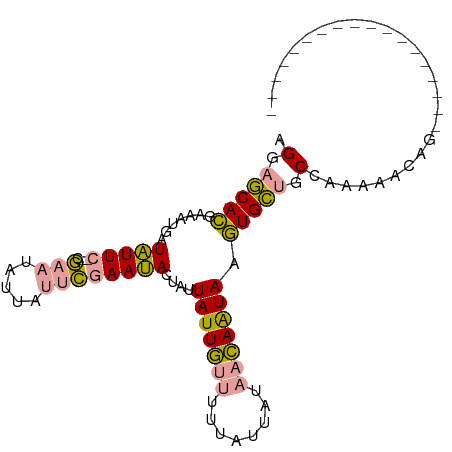
>dm3.chr4 215182 99 + 1351857 AGGAACAAAAGAAGAUUUUAGACAAAUAAAAGGUAAAAAAUAGUAUUAUACAUAAUACA-UAGUAGUUGUGGCAGCACGUUGCAUAUAACUGUGUCUCAA ...................(((((..................((((((....)))))).-...((((((((((((....)))).)))))))))))))... ( -18.10, z-score = -1.77, R) >droSim1.chr4 761064 80 - 949497 AGGAGCACGAAAUGAUAUUC--CGAAUAUUAUUCGAAUACUAUUAUUGUUUUUAUUAUAACAAUAAGUGCUGCCAAGAACAG------------------ .((((((((((.((((((..--...)))))))))........((((((((........))))))))))))).))........------------------ ( -19.20, z-score = -4.01, R) >droSec1.super_52 163193 80 + 192374 AGGAGCACGAAAUGAUAUUC--CGAAUAUUAUUCGAAUACUAUUAUUGUUUUUAUUAUAACAAUAAGUGCUGCCAAAAACAC------------------ .((((((((((.((((((..--...)))))))))........((((((((........))))))))))))).))........------------------ ( -19.20, z-score = -4.25, R) >consensus AGGAGCACGAAAUGAUAUUC__CGAAUAUUAUUCGAAUACUAUUAUUGUUUUUAUUAUAACAAUAAGUGCUGCCAAAAACAG__________________ .(.(((((..((((((((((...))))))))))..........(((((((........))))))).))))).)........................... ( -9.15 = -10.93 + 1.78)

| Location | 215,182 – 215,281 |
|---|---|
| Length | 99 |
| Sequences | 3 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 57.86 |
| Shannon entropy | 0.54179 |
| G+C content | 0.29348 |
| Mean single sequence MFE | -15.73 |
| Consensus MFE | -5.58 |
| Energy contribution | -6.14 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.884173 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
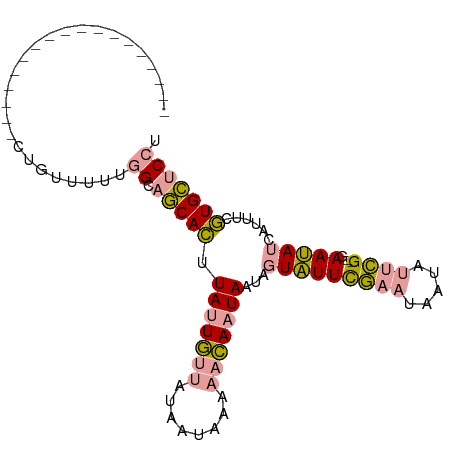
>dm3.chr4 215182 99 - 1351857 UUGAGACACAGUUAUAUGCAACGUGCUGCCACAACUACUA-UGUAUUAUGUAUAAUACUAUUUUUUACCUUUUAUUUGUCUAAAAUCUUCUUUUGUUCCU ...(((((.((((....(((......)))...))))....-.((((((....))))))..................)))))................... ( -10.60, z-score = -0.07, R) >droSim1.chr4 761064 80 + 949497 ------------------CUGUUCUUGGCAGCACUUAUUGUUAUAAUAAAAACAAUAAUAGUAUUCGAAUAAUAUUCG--GAAUAUCAUUUCGUGCUCCU ------------------........((.(((((((((((((........)))))))).....(((((((...)))))--))..........))))))). ( -18.30, z-score = -3.63, R) >droSec1.super_52 163193 80 - 192374 ------------------GUGUUUUUGGCAGCACUUAUUGUUAUAAUAAAAACAAUAAUAGUAUUCGAAUAAUAUUCG--GAAUAUCAUUUCGUGCUCCU ------------------........((.(((((((((((((........)))))))).....(((((((...)))))--))..........))))))). ( -18.30, z-score = -3.32, R) >consensus __________________CUGUUUUUGGCAGCACUUAUUGUUAUAAUAAAAACAAUAAUAGUAUUCGAAUAAUAUUCG__GAAUAUCAUUUCGUGCUCCU ..........................((.(((((.(((((((........)))))))...((((((..............))))))......))))))). ( -5.58 = -6.14 + 0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:01:01 2011