| Sequence ID | dm3.chr4 |
|---|---|
| Location | 181,171 – 181,284 |
| Length | 113 |
| Max. P | 0.669656 |

| Location | 181,171 – 181,284 |
|---|---|
| Length | 113 |
| Sequences | 11 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 64.48 |
| Shannon entropy | 0.79446 |
| G+C content | 0.41133 |
| Mean single sequence MFE | -25.25 |
| Consensus MFE | -7.98 |
| Energy contribution | -8.21 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.32 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.669656 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
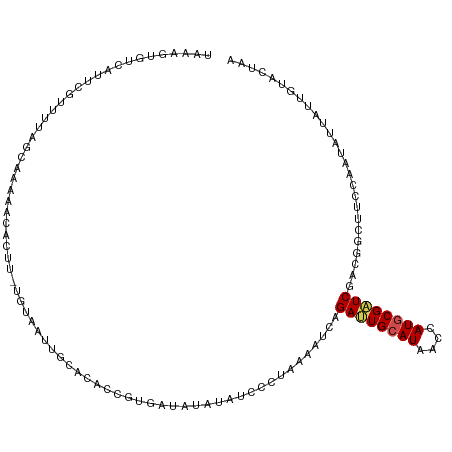
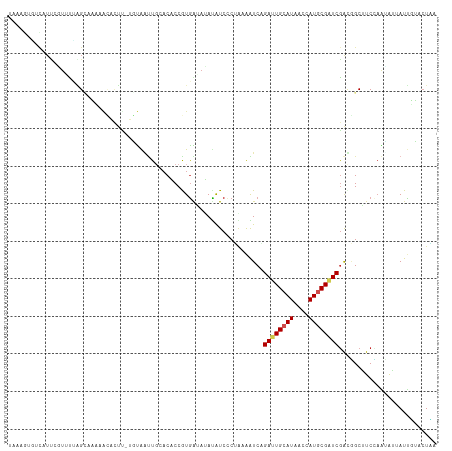
>dm3.chr4 181171 113 + 1351857 UAAAGUGUCAUUCGUUUUAGCAAAAACACUU-UGUAGUUGCACACCGUGAAAUAUAUCCCUGUAAUCAGAUUGCAUAACCAUGCGAUCGGCGGCUUCCAAUAAUAUUUUACUAA ((((((((..((.((....))))..))))))-))............(((((((((.............((((((((....))))))))((......))....)))))))))... ( -22.90, z-score = -1.40, R) >droEre2.scaffold_4512 139715 113 - 1286254 UAACGUGUCAUUCGUUUUAGUCAAAGCAGUU-AGUAGUUGCACACCGUGAAAUAUAUCCCUAUAAUCAGAUUGCAUAACCAUGCGAUCAGCGGCUUCUAACAAUAUUUUACUAU .((((.......)))).((((.(((...(((-((.((((((......(((..((((....)))).)))((((((((....)))))))).)))))).)))))....))).)))). ( -26.10, z-score = -2.83, R) >droYak2.chr4 155701 113 + 1374474 UAACGUUUUCUUGGUUUGAGUCAAAGCUGUU-AGUAGUUGCACACCGUGAAAUAUAUCUCUAUAAUCAGAUUGCAUUACCAUGCGAUCGGCGGCUUCCAACAAUAUUUUACUAA ............(((.((......(((((..-..))))).)).)))(((((((((.............((((((((....))))))))((......))....)))))))))... ( -22.20, z-score = -0.40, R) >droSec1.super_52 123178 113 + 192374 UAAAGUGUCAUUCGUCUUAGCAAACACACUU-UGUAGUUUCACACCGUGAAAUAUAUCCCUAUAAUCAGAUUGCAUAACCAUGCGAUCGGCGGCUUCCAAUAAUAUUUUACUAA ((((((((.....((....))....))))))-))............(((((((((.............((((((((....))))))))((......))....)))))))))... ( -22.00, z-score = -1.97, R) >droSim1.chr4 166784 113 + 949497 UAAAGUGUCAUUCGUCCCAGCAAACACACUU-UGUAGUUUCCCACCGUGAACUAUAUCCCUAUAAUCAGAUUGCAUAACCAUGCGAUCGGCGGCUUCCAAUAAUAUUUUACUAG ((((((((.(((.(((((.............-(((((((..(....)..)))))))............((((((((....)))))))))).)))....))).)))))))).... ( -22.80, z-score = -2.41, R) >droPer1.super_51 475819 113 - 524598 GAUAUUAUCUUUCUUAUUAGCAGCAAUGCUA-AUUAAUUGCACUCCAUGAUAAACGUCCCGAAAGUCCGAUUGCAUAAUCAUGCGAUCGACGGCGUCUAGUAUAAUUCCACUGA .((((((........(((((((....)))))-))...................(((((.(....)..(((((((((....)))))))))..))))).))))))........... ( -26.40, z-score = -2.16, R) >droWil1.scaffold_181130 1037646 108 + 16660200 -----UAACUUCAUUGUCGGUAUCAAUACUC-UUCAAUUGCACACCAUGGUAUAUGUCCCGGAAGUCCGACUGCAUCACCAUUCGAUCGACUGCGUCCAGAAUAAUCGAGCUCA -----.......((((..((((....)))).-..)))).((.....(((((..((((..(((....)))...)))).)))))(((((...(((....)))....)))))))... ( -18.40, z-score = 0.95, R) >droVir3.scaffold_13052 1354033 113 + 2019633 CACUGCGCCAUUCCUGUUGGAGUA-CAAUUUUUUUGAUUGGACACCAUGGUAUACAUCGCGGAAGUCGGACUGCAUGACCAUUCGAUCGAUUGCAUCAAGAAUUAUUGAACUCA ...(((...(((((....))))).-........(((((((((.....((((...(((.((((........))))))))))))))))))))..)))((((......))))..... ( -23.90, z-score = 0.58, R) >droMoj3.scaffold_6498 1743310 113 + 3408170 UACAGCAGCGUCCUUUCGAGAGUU-GAAUUCCUUUGACUGGACCCCGUAGUAGAUAUCACGAAAGUCUGAUUGCAUAAUCAUGCGAUCGAUAGCAUCAACGAUAAUUGAUCUUA (((.((.(.((((..(((((.(..-.....).)))))..)))).).)).)))(.((((((....))..((((((((....)))))))))))).)(((((......))))).... ( -26.50, z-score = -0.97, R) >droGri2.scaffold_14822 374530 113 + 1097346 UACGACGUUAUCGUUGCUCGCGUA-GAAUUCCUUUAAUUGUACGCCGUGAUAAACAUCCCGGAAAUCGGAUUGCAUAACCAUGCGAUCGAUAGCAUCUAGAAUAAUCGAACUCA ...((.((((((((..(..(((((-.((((.....)))).))))).)..)).................((((((((....)))))))))))))).))................. ( -28.20, z-score = -1.29, R) >triCas2.ChLG8 1145904 100 - 15773733 --------------UUCGUGGGUUCGUACGCAUGUACGUGAACCCCAUGGUAGAUAUCCCUCAGAUCUGACUGCAUGACCAUGCGGUCGAGGGCCUCAUGGGUCAUUGCAGUGA --------------.(((((((..(((((....))))).....)))))))(((((.........)))))((((((((((((((.(((.....))).))).))))).)))))).. ( -38.40, z-score = -1.54, R) >consensus UAAAGUGUCAUUCGUUUUAGCAAAAACACUU_UGUAAUUGCACACCGUGAUAUAUAUCCCUAAAAUCAGAUUGCAUAACCAUGCGAUCGACGGCUUCCAAUAUUAUUGUACUAA ....................................................................((((((((....)))))))).......................... ( -7.98 = -8.21 + 0.23)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:00:59 2011