| Sequence ID | dm3.chr4 |
|---|---|
| Location | 124,962 – 125,062 |
| Length | 100 |
| Max. P | 0.556100 |

| Location | 124,962 – 125,062 |
|---|---|
| Length | 100 |
| Sequences | 9 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 77.42 |
| Shannon entropy | 0.46867 |
| G+C content | 0.49471 |
| Mean single sequence MFE | -27.74 |
| Consensus MFE | -18.15 |
| Energy contribution | -18.16 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.36 |
| Mean z-score | -0.91 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.556100 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
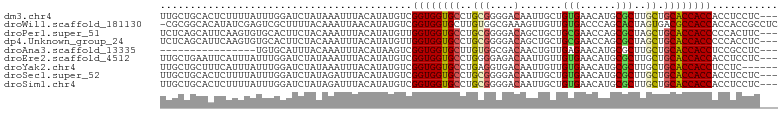
>dm3.chr4 124962 100 - 1351857 UUGCUGCACUCUUUUAUUUGGAUCUAUAAAUUUACAUAUGUCGGUGGUGCCUGCGGGGACAAUUGCUGUGAACAUGCGCUUGCUGCACCACCACCUCCUC--- ....................(((.(((........))).)))((((((((..(((....)....((((....)).))....)).))))))))........--- ( -25.80, z-score = -0.87, R) >droWil1.scaffold_181130 15776334 102 + 16660200 -CGCGGCACAUAUCGAGUCGCUUUUACAAAUUAACAUAUGUCGGUGGUGCUUGUGGCGAAAGUUGUUGUGACCCAGCACUAGUGACGCCACCACCACCGCCUC -.(((((.(.....).)))))..................(.((((((((...((((((..(((.((((.....))))))).....)))))))))))))).).. ( -34.20, z-score = -1.81, R) >droPer1.super_51 192593 100 - 524598 UCUCAGCAUUCAAGUGUGCACUUCUACAAAUUUACAUAUGUUGGUGGUGCCUGCGGGGACAGCUGCUGCGAACCAGCGCUAGCUGCACCACCCCCACUUC--- .....((((......)))).......................((((((((..(((....)(((.((((.....))))))).)).))))))))........--- ( -32.10, z-score = -0.97, R) >dp4.Unknown_group_24 99910 100 - 109830 UCUCAGCAUUCAAGUGUGCACUUCUACAAAUUUACAUAUGUUGGUGGUGCCUGCGGGGACAGCUGCUGCGAACCAGCGCUAGCUGCACCACCCCCACCUC--- .....((((......)))).......................((((((((..(((....)(((.((((.....))))))).)).))))))))........--- ( -32.10, z-score = -0.90, R) >droAna3.scaffold_13335 729250 84 + 3335858 ----------------UGUGCAUUUACAAAUUUACAUAAGUCGGUGGUGCUUGUGGCGACAACUGUUGAGAACAUGCGCUUGCUGCACCACCUCCGCCUC--- ----------------((((.(((....))).))))...(..((((((((....((((.((..(((.....)))))))))....))))))))..).....--- ( -23.90, z-score = -0.77, R) >droEre2.scaffold_4512 86240 100 + 1286254 UUGCUGAAUUCAUUUAUUUGGAUCUAUAAAUUUACAUAUGUCGGUGGUGCCUGGGGAGACAAUUGUUGUGAACAUGCGCUUGCUGCACCACCACCUCCUC--- ...(..(((......)))..).....................((((((((..((.(((.....(((.....)))....))).))))))))))........--- ( -23.20, z-score = -0.09, R) >droYak2.chr4 89481 97 - 1374474 UUGCUGCUUUCAUUUAUUUGGAUCUAUAAAUUUACAUAUGUCGGUGGUGCCUGAGGUGACAAUUGUUGUGAACAUGCGCUUGCUGCACCACCUCCUC------ ........((((......))))....................((((((((..((((((.((..(((.....)))))))))).).)))))))).....------ ( -21.80, z-score = -0.27, R) >droSec1.super_52 67162 100 - 192374 UUGCUGCACUCUUUUAUUUGGAUCUAUAGAUUUACAUAUGUCGGUGGUGCCUGCGGGGACAAUUGCUGUGAACAUGCGCUUGCUGCACCACCACCUCCUC--- ..............(((.((((((....)))))).)))....((((((((..(((....)....((((....)).))....)).))))))))........--- ( -28.30, z-score = -1.27, R) >droSim1.chr4 132465 100 - 949497 UUGCUGCACUCUUUUAUUUGGAUCUAUAGAUUUACAUAUGUCGGUGGUGCCUGCGGGGACAAUUGCUGUGAACAUGCGCUUGCUGCACCACCACCUCCUC--- ..............(((.((((((....)))))).)))....((((((((..(((....)....((((....)).))....)).))))))))........--- ( -28.30, z-score = -1.27, R) >consensus UUGCUGCACUCAUUUAUUUGGAUCUAUAAAUUUACAUAUGUCGGUGGUGCCUGCGGGGACAAUUGCUGUGAACAUGCGCUUGCUGCACCACCACCUCCUC___ ..........................................((((((((..(((....).......(((......)))..)).))))))))........... (-18.15 = -18.16 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:00:58 2011