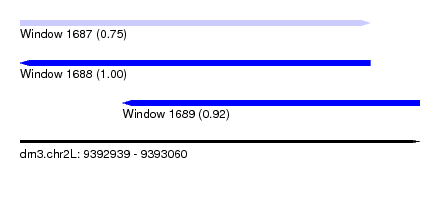
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 9,392,939 – 9,393,060 |
| Length | 121 |
| Max. P | 0.997563 |

| Location | 9,392,939 – 9,393,045 |
|---|---|
| Length | 106 |
| Sequences | 8 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 66.76 |
| Shannon entropy | 0.64384 |
| G+C content | 0.56174 |
| Mean single sequence MFE | -31.79 |
| Consensus MFE | -17.36 |
| Energy contribution | -17.99 |
| Covariance contribution | 0.63 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.65 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.753971 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
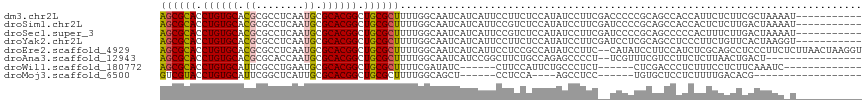
>dm3.chr2L 9392939 106 + 23011544 -----------AUUUUAGCGAAGAGAAUGGUGGCUGCGGGGGUCGAAGGAUAUGGAGAAGGAAUGAUGAUUGCCAAAAGCGCAGCCGUGCGCAUUGAGGCGCGUGCACAGGUGCGCU -----------....................(((((((...(((....)))........(((((....))).)).....)))))))..((((......))))(((((....))))). ( -32.80, z-score = -0.27, R) >droSim1.chr2L 9171984 106 + 22036055 -----------AUUUUAGUCAAGAGAGUGGUGGCUGCGGGGAUCGAAGGAUAUGGAGACGGAAUGAUGAUUGCCAAAAGCGCAGCCGUGCGCAUUGAGGCGCGUGCACAGGUGCGCU -----------...(((.((....)).))).(((((((...(((....)))..(....)(((((....))).)).....)))))))..((((......))))(((((....))))). ( -34.20, z-score = -0.44, R) >droSec1.super_3 4849739 106 + 7220098 -----------AUUUUAGUCAAGAAAGUGGGGGCUGCGGGGAUCGAAGGAUAUGGAGACGGAAUGAUGAUUGCCAAAAGCGCAGCCGUGCGCAUUGAGGCGCGUGCACAGGUGCGCU -----------....................(((((((...(((....)))..(....)(((((....))).)).....)))))))..((((......))))(((((....))))). ( -33.70, z-score = -0.51, R) >droYak2.chr2L 12066767 106 + 22324452 -----------ACCUUAGUGAACAGAAGGGAGGCUGCGAGGAUCGAAGGAUAUGGAGAAGGAAUGAUGAUUGCCAAAAGCGCAGCCGUGCGCAUUGAGGCGCGUGCACAGGUGCGCU -----------.((((.(....)..))))..(((((((...(((....)))........(((((....))).)).....)))))))..((((......))))(((((....))))). ( -36.00, z-score = -1.71, R) >droEre2.scaffold_4929 10005066 115 + 26641161 ACCUUAGUUAAGAGAAGGGAGGCUGCGAGAUGGAAGGAUAUG--GAAGGAUAUGGCGGAGGAAUGAUGAUUGCCAAAAGCGCAGCCGUGCGCAUUGAGGCGCGUGCACAGGUGCGCU .((((.........))))..(((((((..........((((.--.....))))(((((...........))))).....)))))))..((((......))))(((((....))))). ( -34.20, z-score = -0.91, R) >droAna3.scaffold_12943 745790 99 + 5039921 ----------------AGUCAGUUAAGAGAAGGACGAAACGA--AGGGGCUCUGGCAGAAGCCGGAUGAUUGCCAAAAGCGCAGCCGUGCGCAUUGGUGCGCGUGCACAGGUGCGCU ----------------.(((..((....))..)))....(..--..)(((((((((....)))))).....)))...(((((((((((((((....))))))).)).....)))))) ( -36.60, z-score = -0.86, R) >droWil1.scaffold_180772 1320823 92 - 8906247 -------------GAUUUGAAGAGGAAAGAGGGUCGAG------AGAGGGCAGAAUGGAAG------GAUAUCGAAAAGCGCAGCCGUGCGCAUUCAGGCGAAUGCACAGGUGCGCU -------------....................((((.------......(......)...------....))))..((((((.((.((.((((((....)))))).)))))))))) ( -23.84, z-score = -0.79, R) >droMoj3.scaffold_6500 16278430 83 + 32352404 ------------------CGUGUCAAAAGAGGAGCACA------GGAGGCU----UGGAGG------AGCUGCCAAAAGCGCAGCCGUGCGCAAUGAGCCGAAUGCACAGGUACGAC ------------------.((((.......((..((..------((.((((----(....)------)))).))....(((((....)))))..))..))....))))......... ( -23.00, z-score = 0.26, R) >consensus ___________A_UUUAGUCAAGAGAAAGAGGGCUGCGGGGA__GAAGGAUAUGGAGAAGGAAUGAUGAUUGCCAAAAGCGCAGCCGUGCGCAUUGAGGCGCGUGCACAGGUGCGCU .............................................................................((((((.(.((((((..........)))))).).)))))) (-17.36 = -17.99 + 0.63)

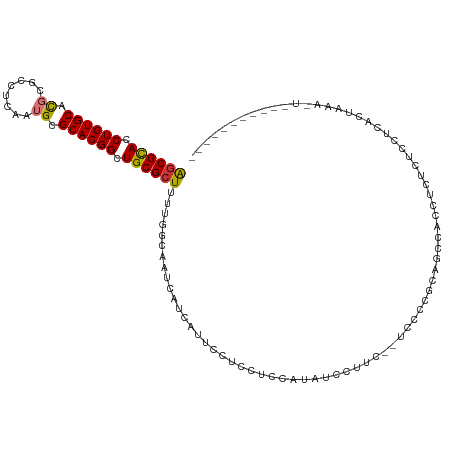
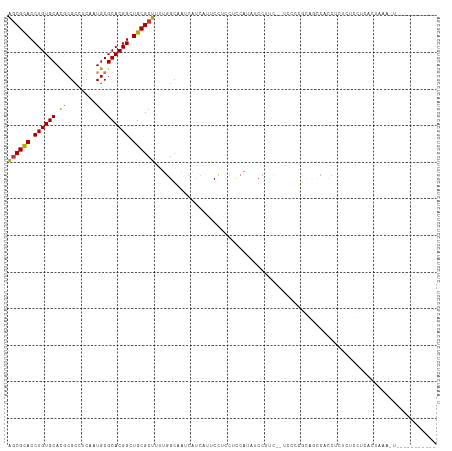
| Location | 9,392,939 – 9,393,045 |
|---|---|
| Length | 106 |
| Sequences | 8 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 66.76 |
| Shannon entropy | 0.64384 |
| G+C content | 0.56174 |
| Mean single sequence MFE | -25.75 |
| Consensus MFE | -17.74 |
| Energy contribution | -17.71 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.13 |
| SVM RNA-class probability | 0.997563 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 9392939 106 - 23011544 AGCGCACCUGUGCACGCGCCUCAAUGCGCACGGCUGCGCUUUUGGCAAUCAUCAUUCCUUCUCCAUAUCCUUCGACCCCCGCAGCCACCAUUCUCUUCGCUAAAAU----------- ((((((....)))..((((......))))..((((((((.....)).....................((....)).....))))))............))).....----------- ( -26.00, z-score = -1.98, R) >droSim1.chr2L 9171984 106 - 22036055 AGCGCACCUGUGCACGCGCCUCAAUGCGCACGGCUGCGCUUUUGGCAAUCAUCAUUCCGUCUCCAUAUCCUUCGAUCCCCGCAGCCACCACUCUCUUGACUAAAAU----------- .((((....))))..((((......))))..((((((((.....))....................(((....)))....))))))....................----------- ( -25.90, z-score = -1.52, R) >droSec1.super_3 4849739 106 - 7220098 AGCGCACCUGUGCACGCGCCUCAAUGCGCACGGCUGCGCUUUUGGCAAUCAUCAUUCCGUCUCCAUAUCCUUCGAUCCCCGCAGCCCCCACUUUCUUGACUAAAAU----------- .((((....))))..((((......))))..((((((((.....))....................(((....)))....))))))....................----------- ( -25.00, z-score = -1.29, R) >droYak2.chr2L 12066767 106 - 22324452 AGCGCACCUGUGCACGCGCCUCAAUGCGCACGGCUGCGCUUUUGGCAAUCAUCAUUCCUUCUCCAUAUCCUUCGAUCCUCGCAGCCUCCCUUCUGUUCACUAAGGU----------- .((((....))))..((((......))))..((((((((.....))....................(((....)))....))))))..((((..(....).)))).----------- ( -26.80, z-score = -1.34, R) >droEre2.scaffold_4929 10005066 115 - 26641161 AGCGCACCUGUGCACGCGCCUCAAUGCGCACGGCUGCGCUUUUGGCAAUCAUCAUUCCUCCGCCAUAUCCUUC--CAUAUCCUUCCAUCUCGCAGCCUCCCUUCUCUUAACUAAGGU .((((....))))..((((......))))..(((((((....((((...............))))(((.....--.)))...........)))))))..((((.........)))). ( -28.06, z-score = -2.45, R) >droAna3.scaffold_12943 745790 99 - 5039921 AGCGCACCUGUGCACGCGCACCAAUGCGCACGGCUGCGCUUUUGGCAAUCAUCCGGCUUCUGCCAGAGCCCCU--UCGUUUCGUCCUUCUCUUAACUGACU---------------- ((((((.((((((..(((......))))))))).))))))...(((.....((.(((....))).)))))...--.......(((............))).---------------- ( -28.10, z-score = -0.47, R) >droWil1.scaffold_180772 1320823 92 + 8906247 AGCGCACCUGUGCAUUCGCCUGAAUGCGCACGGCUGCGCUUUUCGAUAUC------CUUCCAUUCUGCCCUCU------CUCGACCCUCUUUCCUCUUCAAAUC------------- (((((((((((((((((....))))))))).)).))))))..((((....------.................------.))))....................------------- ( -24.75, z-score = -4.34, R) >droMoj3.scaffold_6500 16278430 83 - 32352404 GUCGUACCUGUGCAUUCGGCUCAUUGCGCACGGCUGCGCUUUUGGCAGCU------CCUCCA----AGCCUCC------UGUGCUCCUCUUUUGACACG------------------ ((((.......((((..((((...((.(...((((((.(....)))))))------...)))----))))...------.))))........))))...------------------ ( -21.36, z-score = -0.18, R) >consensus AGCGCACCUGUGCACGCGCCUCAAUGCGCACGGCUGCGCUUUUGGCAAUCAUCAUUCCUCCUCCAUAUCCUUC__UCCCCGCAGCCACCUCUCUCCUCACUAAA_U___________ ((((((.((((((.((........)).)))))).))))))............................................................................. (-17.74 = -17.71 + -0.03)
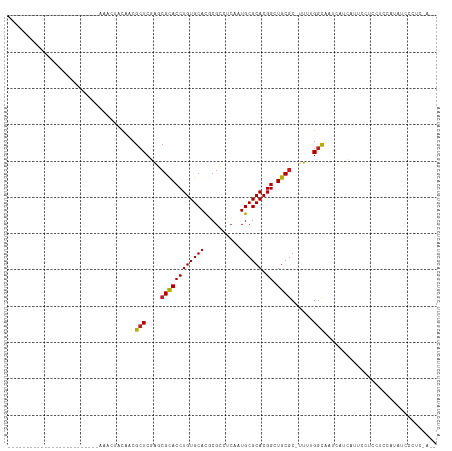
| Location | 9,392,970 – 9,393,060 |
|---|---|
| Length | 90 |
| Sequences | 12 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 70.83 |
| Shannon entropy | 0.54360 |
| G+C content | 0.56053 |
| Mean single sequence MFE | -25.63 |
| Consensus MFE | -15.23 |
| Energy contribution | -14.95 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.917016 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
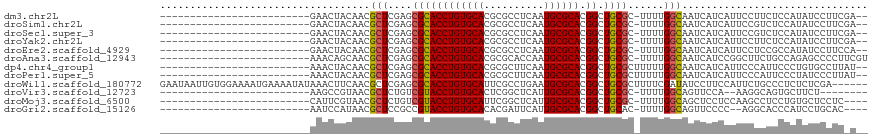
>dm3.chr2L 9392970 90 - 23011544 -------------------------GAACUACAACGCUCGAGCGCACCUGUGCACGCGCCUCAAUGCGCACGGCUGCGC-UUUUGGCAAUCAUCAUUCCUUCUCCAUAUCCUUCGA-- -------------------------..........(((.(((((((.((((((..(((......))))))))).)))))-))..))).............................-- ( -25.30, z-score = -1.95, R) >droSim1.chr2L 9172015 90 - 22036055 -------------------------GAACUACAACGCUCGAGCGCACCUGUGCACGCGCCUCAAUGCGCACGGCUGCGC-UUUUGGCAAUCAUCAUUCCGUCUCCAUAUCCUUCGA-- -------------------------..........(((.(((((((.((((((..(((......))))))))).)))))-))..))).............................-- ( -25.30, z-score = -1.30, R) >droSec1.super_3 4849770 90 - 7220098 -------------------------GAACUACAACGCUCGAGCGCACCUGUGCACGCGCCUCAAUGCGCACGGCUGCGC-UUUUGGCAAUCAUCAUUCCGUCUCCAUAUCCUUCGA-- -------------------------..........(((.(((((((.((((((..(((......))))))))).)))))-))..))).............................-- ( -25.30, z-score = -1.30, R) >droYak2.chr2L 12066798 90 - 22324452 -------------------------GAACUACAACGCUCGAGCGCACCUGUGCACGCGCCUCAAUGCGCACGGCUGCGC-UUUUGGCAAUCAUCAUUCCUUCUCCAUAUCCUUCGA-- -------------------------..........(((.(((((((.((((((..(((......))))))))).)))))-))..))).............................-- ( -25.30, z-score = -1.95, R) >droEre2.scaffold_4929 10005106 90 - 26641161 -------------------------GAACUACAACGCUCGAGCGCACCUGUGCACGCGCCUCAAUGCGCACGGCUGCGC-UUUUGGCAAUCAUCAUUCCUCCGCCAUAUCCUUCCA-- -------------------------..........(((.(((((((.((((((..(((......))))))))).)))))-))..))).............................-- ( -25.30, z-score = -1.76, R) >droAna3.scaffold_12943 745812 92 - 5039921 -------------------------AAACAGCAACGCUCGAGCGCACCUGUGCACGCGCACCAAUGCGCACGGCUGCGC-UUUUGGCAAUCAUCCGGCUUCUGCCAGAGCCCCUUCGU -------------------------..........(((((((((((.((((((..(((......))))))))).)))))-)).(((((.............)))))))))........ ( -32.82, z-score = -0.83, R) >dp4.chr4_group1 4370826 91 + 5278887 -------------------------AAACUACAACGCUCGAGCGCACCUGUGCACGCGCUUCAAUGCGCACGGCUGCGCUUUUUGGCAAUCAUCAUUCCCAUUCCCUGUGCCUUAU-- -------------------------..........(((.(((((((.((((((..(((......))))))))).)))))))...))).............................-- ( -24.90, z-score = -1.31, R) >droPer1.super_5 4301915 91 - 6813705 -------------------------AAACUACAACGCUCGAGCGCACCUGUGCACGCGCUUCAAUGCGCACGGCUGCGCUUUUUGGCAAUCAUCAUUCCCAUUCCCUAUCCCUUAU-- -------------------------..........(((.(((((((.((((((..(((......))))))))).)))))))...))).............................-- ( -24.90, z-score = -2.65, R) >droWil1.scaffold_180772 1320843 112 + 8906247 GAAUAAUUGUGGAAAAUGAAAAUAUAAACUUCAACGCUCGAGCGCACCUGUGCAUUCGCCUGAAUGCGCACGGCUGCGCUUUUCGAUAUCCUUCCAUUCUGCCCUCUCUCGA------ ........((((((..((((.........)))).((...((((((((((((((((((....))))))))).)).)))))))..))......))))))...............------ ( -32.90, z-score = -3.49, R) >droVir3.scaffold_12723 416510 82 + 5802038 -------------------------AAGCCGUAACGCUCUGUCGUACCUGUGCACUCGGCUCAUUGCGCACGGCUGCGC-UUUUGGCAGUUCCA--AAGGCAGUGCUUCU-------- -------------------------.((((((..(((...((((............)))).....))).))))))..((-((((((.....)))--))))).........-------- ( -26.10, z-score = 0.06, R) >droMoj3.scaffold_6500 16278440 88 - 32352404 -------------------------CAUUCGUAACGCUCUGUCGUACCUGUGCAUUCGGCUCAUUGCGCACGGCUGCGC-UUUUGGCAGCUCCUCCAAGCCUCCUGUGCUCCUC---- -------------------------..........(((.((.((((..((.((.....)).)).)))))).))).((((-....(((...........)))....)))).....---- ( -20.00, z-score = 0.70, R) >droGri2.scaffold_15126 6241684 86 + 8399593 -------------------------AAUCCAUAACGCUCCGCCGUACCUGUGCACACGAUUCAUUGCGCACGGCUGCAC-UUUUGGCAGUUCCCC--AGGCACCCAUCCUGCAC---- -------------------------...............((((((.((((((.((........)).)))))).)))..-...(((.......))--)))).............---- ( -19.40, z-score = 0.06, R) >consensus _________________________AAACUACAACGCUCGAGCGCACCUGUGCACGCGCCUCAAUGCGCACGGCUGCGC_UUUUGGCAAUCAUCAUUCCUCCUCCAUAUCCCUC_A__ ...................................(((..(.((((((((((((..........)))))).)).))))...)..)))............................... (-15.23 = -14.95 + -0.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:28:09 2011