| Sequence ID | dm3.chr3R |
|---|---|
| Location | 27,742,104 – 27,742,210 |
| Length | 106 |
| Max. P | 0.936253 |

| Location | 27,742,104 – 27,742,196 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 85.22 |
| Shannon entropy | 0.29036 |
| G+C content | 0.46300 |
| Mean single sequence MFE | -24.47 |
| Consensus MFE | -18.30 |
| Energy contribution | -19.55 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.936253 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
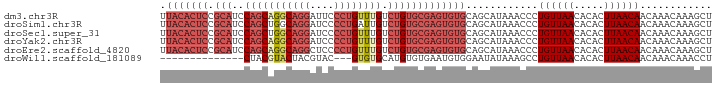
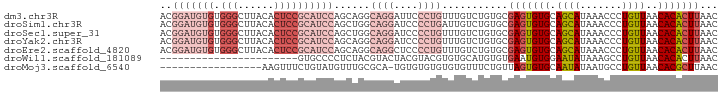
>dm3.chr3R 27742104 92 - 27905053 UUACACUCCGCAUCCAGCAGGCAGGAUUCCCUGUUUGUCUGUGCGAGUGUGCAGCAUAAACCCUGUUAACACACUUAACAACAAACAAAGCU .(((((((.(((..(((((((((((....)))))))).))))))))))))).(((........((((((.....)))))).........))) ( -28.93, z-score = -3.33, R) >droSim1.chr3R 27357063 92 - 27517382 UUACACUCCGCAUCCAGCUGGCAGGAUCCCCUGAUUGUCUGUGCGAGUGUGCAGCAUAAACCCUGUUAACACACUUAACAACAAACAAAGCU .(((((((((((..(((....((((....)))).)))..)))).))))))).(((........((((((.....)))))).........))) ( -23.33, z-score = -1.63, R) >droSec1.super_31 396843 92 - 568655 UUACACUCCGCAUCCAGCUGGCAGGAUCCCCUGUUUGUCUGUGCGAGUGUGCAGCAUAAACCCUGUUAACACACUUAACAACAAACAAAGCU .(((((((.(((..((((..(((((....)))))..).))))))))))))).(((........((((((.....)))))).........))) ( -25.53, z-score = -2.16, R) >droYak2.chr3R 28671772 92 - 28832112 UUACACUCCGCAUCCAGCAGGCAGGAUCCCCUGUUUGUCUGUGCGAGUGUGCAGCAUAAACCCUGUUAACACACUUAACAACAAACAAAGCU .(((((((.(((..(((((((((((....)))))))).))))))))))))).(((........((((((.....)))))).........))) ( -28.93, z-score = -3.41, R) >droEre2.scaffold_4820 10245604 92 + 10470090 UUACACUCCGCAUCCAGCAGGCAGGCUCCCCUGUUUGUCUGUGCGAGUGUGCAGCAUAAACCCUGUUAACACACUUAACAACAAACAAAGCU .(((((((.(((..(((((((((((....)))))))).))))))))))))).(((........((((((.....)))))).........))) ( -28.13, z-score = -2.82, R) >droWil1.scaffold_181089 10301033 75 - 12369635 --------------CUACGUACUACGUAC---GUGUGCAUGUGUGAAUGUGGAAUAUAAAGCCUGUUAACACACUUAACAACAAACAAACCU --------------..((((((...))))---))((....(((((...(..(..........)..)...)))))...))............. ( -12.00, z-score = 0.67, R) >consensus UUACACUCCGCAUCCAGCAGGCAGGAUCCCCUGUUUGUCUGUGCGAGUGUGCAGCAUAAACCCUGUUAACACACUUAACAACAAACAAAGCU .........((((...(((((((((....)))))))))..))))(((((((.((((.......))))..)))))))................ (-18.30 = -19.55 + 1.25)
| Location | 27,742,117 – 27,742,210 |
|---|---|
| Length | 93 |
| Sequences | 7 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 71.69 |
| Shannon entropy | 0.54481 |
| G+C content | 0.49485 |
| Mean single sequence MFE | -27.96 |
| Consensus MFE | -12.36 |
| Energy contribution | -15.69 |
| Covariance contribution | 3.33 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.612887 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
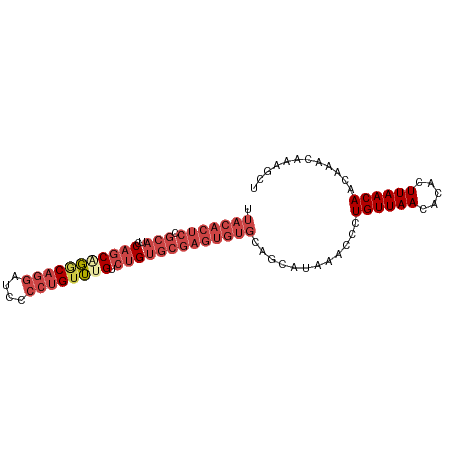
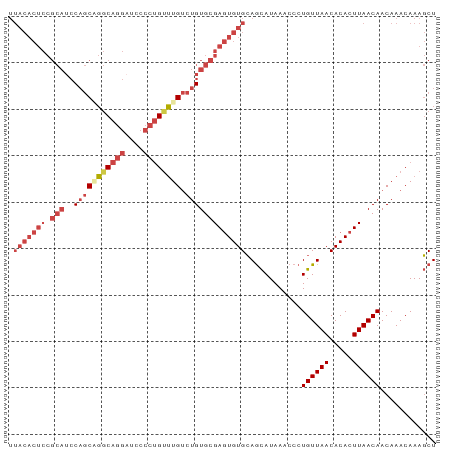
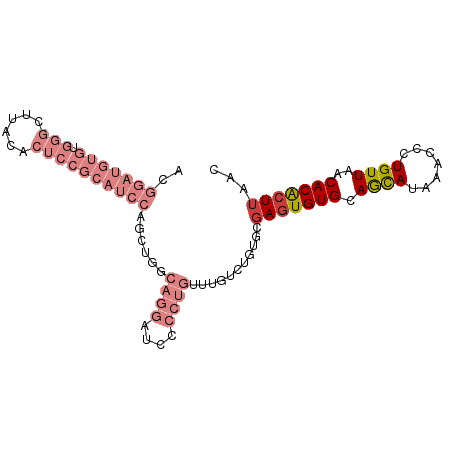
>dm3.chr3R 27742117 93 - 27905053 ACGGAUGUGUGGGCUUACACUCCGCAUCCAGCAGGCAGGAUUCCCUGUUUGUCUGUGCGAGUGUGCAGCAUAAACCCUGUUAACACACUUAAC ......(((((.((((((((((.(((..(((((((((((....)))))))).))))))))))))).)))...(((...)))..)))))..... ( -35.30, z-score = -2.74, R) >droSim1.chr3R 27357076 93 - 27517382 ACGGAUGUGUGGGCUUACACUCCGCAUCCAGCUGGCAGGAUCCCCUGAUUGUCUGUGCGAGUGUGCAGCAUAAACCCUGUUAACACACUUAAC ......(((((.((((((((((((((..(((....((((....)))).)))..)))).))))))).)))...(((...)))..)))))..... ( -29.70, z-score = -1.11, R) >droSec1.super_31 396856 93 - 568655 ACGGAUGUGUGGGCUUACACUCCGCAUCCAGCUGGCAGGAUCCCCUGUUUGUCUGUGCGAGUGUGCAGCAUAAACCCUGUUAACACACUUAAC ......(((((.((((((((((.(((..((((..(((((....)))))..).))))))))))))).)))...(((...)))..)))))..... ( -31.90, z-score = -1.65, R) >droYak2.chr3R 28671785 93 - 28832112 ACGGAUGUGUGGGCUUACACUCCGCAUCCAGCAGGCAGGAUCCCCUGUUUGUCUGUGCGAGUGUGCAGCAUAAACCCUGUUAACACACUUAAC ......(((((.((((((((((.(((..(((((((((((....)))))))).))))))))))))).)))...(((...)))..)))))..... ( -35.30, z-score = -2.64, R) >droEre2.scaffold_4820 10245617 93 + 10470090 ACGGAUGUGUGGGCUUACACUCCGCAUCCAGCAGGCAGGCUCCCCUGUUUGUCUGUGCGAGUGUGCAGCAUAAACCCUGUUAACACACUUAAC ..(((((((.(((......)))))))))).((((((((((......))))))))))..(((((((.((((.......))))..)))))))... ( -36.10, z-score = -2.63, R) >droWil1.scaffold_181089 10301046 70 - 12369635 -----------------------GUGCCCCUCUACGUACUACGUACGUGUGCAUGUGUGAAUGUGGAAUAUAAAGCCUGUUAACACACUUAAC -----------------------((((.....(((((((...))))))).))))(((((...(..(..........)..)...)))))..... ( -13.90, z-score = 0.00, R) >droMoj3.scaffold_6540 18389031 75 - 34148556 -----------------AAGUUUCUGUAUGUUUGCGCA-UGUGUGUGUGUGUUUCUGUUAGUGUGCAAUAUAAUGCCUGUUAACACGCUUAAC -----------------..(((...(..((((.(((((-(.....((((..((......))..)))).....))))..)).))))..)..))) ( -13.50, z-score = 0.67, R) >consensus ACGGAUGUGUGGGCUUACACUCCGCAUCCAGCUGGCAGGAUCCCCUGUUUGUCUGUGCGAGUGUGCAGCAUAAACCCUGUUAACACACUUAAC ..(((((((.(((......))))))))))......((((....))))...........(((((((.((((.......))))..)))))))... (-12.36 = -15.69 + 3.33)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:00:49 2011