| Sequence ID | dm3.chr3R |
|---|---|
| Location | 27,665,635 – 27,665,725 |
| Length | 90 |
| Max. P | 0.932104 |

| Location | 27,665,635 – 27,665,725 |
|---|---|
| Length | 90 |
| Sequences | 8 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 80.25 |
| Shannon entropy | 0.40404 |
| G+C content | 0.64723 |
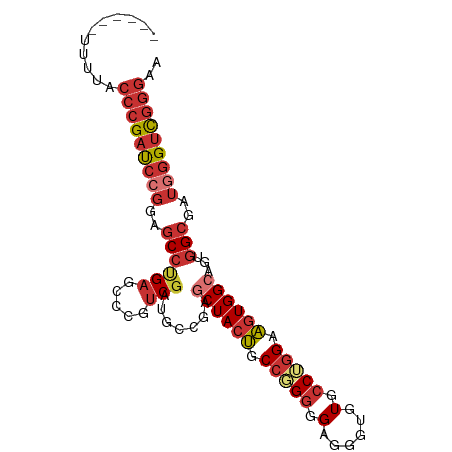
| Mean single sequence MFE | -40.46 |
| Consensus MFE | -29.43 |
| Energy contribution | -30.84 |
| Covariance contribution | 1.41 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.37 |
| SVM RNA-class probability | 0.932104 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
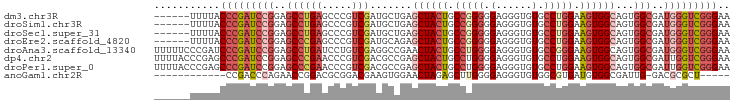
>dm3.chr3R 27665635 90 - 27905053 ------UUUUACCCGAUCCGGAGCCUGAGCCCGUCGAUGCUGAGCUACUGCCGGGGGAGGGUGUGCCUGGAAGUGGCAGUGGCGAUGGGUCGGGAA ------.....(((((((((..((((.(((........))).)((((((.(((((.(......).))))).))))))...)))..))))))))).. ( -43.20, z-score = -2.49, R) >droSim1.chr3R 27288719 90 - 27517382 ------UUUUACCCGAUCCGGAGCCUGAGCCCGUCGAUGCUGAGCUACUGCCGGGGGAGGGUGUGCCUGGAAGUGGCAGUGGCGAUGGGUCGGGAA ------.....(((((((((..((((.(((........))).)((((((.(((((.(......).))))).))))))...)))..))))))))).. ( -43.20, z-score = -2.49, R) >droSec1.super_31 329046 90 - 568655 ------UUUUACCCGAUCCGGAGCCUGAGCCCGUCGAUGCUGAGCUACUGCCGGGGGAGGGUGUGCCUGGAAGUGGCAGUGGCGAUGGGUCGGGAA ------.....(((((((((..((((.(((........))).)((((((.(((((.(......).))))).))))))...)))..))))))))).. ( -43.20, z-score = -2.49, R) >droEre2.scaffold_4820 10167268 90 + 10470090 ------UUUUACCCGAUCCGGAGCCCGAGCCCGUCGAUGCAGAGCUACUGCCGGGGGAGGGUGUGCCUGGAAGUGGCAGUGGCGAUGGGUCGGGAA ------.....(((((((((..((((..((........))...((((((.(((((.(......).))))).)))))).).)))..))))))))).. ( -42.40, z-score = -1.97, R) >droAna3.scaffold_13340 23416073 96 + 23697760 UUUUUCCCGAUCCCGAUCCGGAGCCUGAUCCUGUCGAGGCCGAACUACUGCCUGGGGAGGGUGUGCCGGGAAGUGGCAGUGGCGAUGGGUCGGGAA ..........((((((((((..(((..(((((.((.((((.(.....).)))).)).))))).(((((.....)))))..)))..)))))))))). ( -46.60, z-score = -2.28, R) >dp4.chr2 14665070 96 + 30794189 UUUUACCCGAGCCCGAUCCGGAGCCCGAACCCGUCGACGCCGAGCUACUGCCUGGGGAGGGUGUGCCUGGAAGUGGCAGUGGCGAUUGGUCGGGAA .....(((((..(((((((((.(..(((.....))).).))).(((((((((......(((....)))......)))))))))))))))))))).. ( -42.60, z-score = -1.36, R) >droPer1.super_0 4276715 96 - 11822988 UUUUACCCGAGCCCGAUCCGGAGCCCGAACCCGUCGACGCCGAGCUACUGCCUGGGGAGGGUGUGCCUGGAAGUGGCAGUGGCGAUUGGUCGGGAA .....(((((..(((((((((.(..(((.....))).).))).(((((((((......(((....)))......)))))))))))))))))))).. ( -42.60, z-score = -1.36, R) >anoGam1.chr2R 48478508 78 - 62725911 ------------CCGACCCAGAACCGGACGCGGACGAAGUGGAACUAGAGCUUGGGGAGGGUGUGGCGUGAUGUGGCGAUUG-GACGCGCU----- ------------((..(((....(((....)))...((((.........)))))))..))(((((.(((......)))....-..))))).----- ( -19.90, z-score = 1.31, R) >consensus ______UUUUACCCGAUCCGGAGCCUGAGCCCGUCGAUGCCGAGCUACUGCCGGGGGAGGGUGUGCCUGGAAGUGGCAGUGGCGAUGGGUCGGGAA ...........(((((((((..((((((.....))).......((((((.(((((.(......).))))).))))))...)))..))))))))).. (-29.43 = -30.84 + 1.41)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:00:44 2011