
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 27,602,727 – 27,602,828 |
| Length | 101 |
| Max. P | 0.670287 |

| Location | 27,602,727 – 27,602,828 |
|---|---|
| Length | 101 |
| Sequences | 10 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 63.17 |
| Shannon entropy | 0.78113 |
| G+C content | 0.37364 |
| Mean single sequence MFE | -18.35 |
| Consensus MFE | -4.70 |
| Energy contribution | -3.82 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 2.55 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.26 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.670287 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 27602727 101 + 27905053 ----ACCGCGCGAACCGGUUCAAUUCAUAACUAGUUCG--CCCGAACUGCACAACCC--UAGCAAAAGAAAUAGCUUAAAUAUAAUUUAUUUAACGCGA-AUUGGACAAC ----.............(((((((((.....((((((.--...))))))........--..............(((((((((.....))))))).))))-)))))))... ( -22.10, z-score = -2.76, R) >droAna3.scaffold_12911 1881151 99 - 5364042 AGCCGGGAAGCAAUUUUUAUCGGGUCAUAAGCGGUCAC----ACUGCUGCACAACCC------AAAAGAAAUAACUAAAAUAUAAUUAA-UUAACGAAAUAUUGCACUAA .........(((((.(((...((((....(((((....----.))))).....))))------...((......)).............-......))).)))))..... ( -16.90, z-score = -0.89, R) >droEre2.scaffold_4820 10102029 101 - 10470090 ----ACCGCGCGAACCGGUUCAGUUCAAAACUAGUUCG--CCCGAACUGCACAACCC--UAGCAAAAGAAAUAGCUUAAAUAUAAUUUAUUUAACGCGA-AUUGGACAGC ----..((.((((((..(((........)))..)))))--).))..(((..(((..(--(......)).....(((((((((.....))))))).))..-.)))..))). ( -22.50, z-score = -1.96, R) >droYak2.chr3R 28537140 101 + 28832112 ----ACCGCGCGAACCGGUUCAGUUCAAAACUAGUUCG--CCCGAACUGCACAACCC--UAGCAAAAGAAAUAGCUUAAAUAUAAUUUAUUUAACGCGA-AUUGGACAGC ----..((.((((((..(((........)))..)))))--).))..(((..(((..(--(......)).....(((((((((.....))))))).))..-.)))..))). ( -22.50, z-score = -1.96, R) >droSec1.super_31 257220 101 + 568655 ----ACCGAGCGAACCGGUUCAGUUCACAACUAGUUCG--CCCGAACUGCACAACCC--UAGCAAAAGAAAUAUCUUAAAUAUAAUUUAUUUAACGCGA-AUUGGACAAC ----...(((((((....))).))))....((((((((--(......(((.......--..)))...........(((((((.....))))))).))))-)))))..... ( -20.60, z-score = -2.47, R) >droSim1.chr3R 27220454 101 + 27517382 ----ACCGCGCGAACCGGUUCAGUUCACAACUAGUUCG--CCCGAACUGCACAACCC--UAGCAAAAGAAAUAGCUUAAAUAUAAUUUAUUUAACGCGA-AUUGGACAAC ----.............(((((((((.....((((((.--...))))))........--..............(((((((((.....))))))).))))-)))))))... ( -22.10, z-score = -2.34, R) >droWil1.scaffold_181009 1696765 88 + 3585778 -----------------ACUUAGGUAGGAACACGUUUU----GCGAUUUUAGAAUUUGUUUGUCAAAUAAACAUUCUGCAUAUAAUUAAUUAAACACACUGUGGAAGAC- -----------------.(((..((((((...((....----.))...........(((((((...)))))))))))))...............(((...))).)))..- ( -10.30, z-score = 0.97, R) >droPer1.super_0 5428926 99 - 11822988 ----ACUAAUUGAACUUGUUCAGUUCA-AACUAGUUCAGACCUGAACUGCACAACUC--UAGCAAAGAAAAUAAAUAAAAUACA-CUAAUCUAACGAAAUAUUGCAC--- ----.....(((((((.....))))))-)...((((((....))))))(((....((--(.....)))................-....((....)).....)))..--- ( -14.80, z-score = -1.83, R) >dp4.chr2 14047790 99 + 30794189 ----ACUAAUUGAACUUGUUCAGUUCA-AACUAGUUCAGACCUGAACUGCACAACUC--UAGCAAAGAAAAUAAAUAAAAUACA-CUAAUUUAACGAAAUAUUGCAC--- ----.....(((((((.....))))))-)..(((((((....)))))))........--..((((......(((((........-...)))))........))))..--- ( -13.24, z-score = -1.14, R) >droVir3.scaffold_13047 11135380 89 + 19223366 ---AGGUCGACAAUCGACCGGCUACUAUAAUCUAUCGG----CCGUCUACACAAGU-------AAAAAUAAUAAUUAAC-UCUAAUAUA-CGAAUUGGAAGUAGC----- ---.(((((.....))))).((((((............----.....(((....))-------)...............-((((((...-...))))))))))))----- ( -18.50, z-score = -2.01, R) >consensus ____ACCGAGCGAACCGGUUCAGUUCAUAACUAGUUCG__CCCGAACUGCACAACCC__UAGCAAAAGAAAUAACUUAAAUAUAAUUUAUUUAACGCGA_AUUGGACAAC ....((((.......))))............(((((((....)))))))............................................................. ( -4.70 = -3.82 + -0.88)
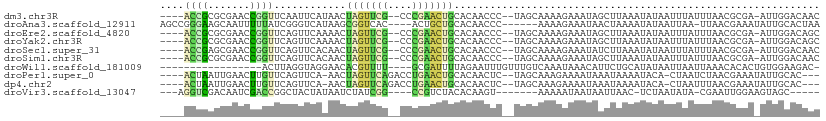
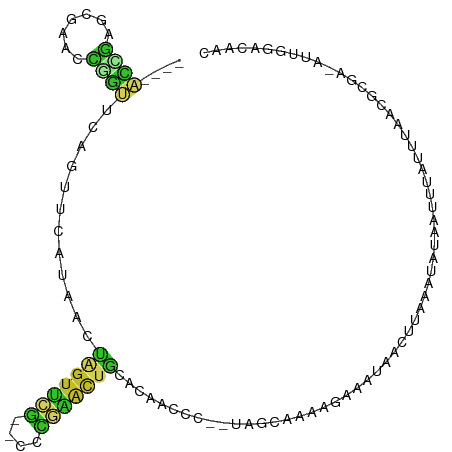

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:00:40 2011