| Sequence ID | dm3.chr3R |
|---|---|
| Location | 27,601,339 – 27,601,439 |
| Length | 100 |
| Max. P | 0.783895 |

| Location | 27,601,339 – 27,601,439 |
|---|---|
| Length | 100 |
| Sequences | 14 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 66.20 |
| Shannon entropy | 0.75183 |
| G+C content | 0.41544 |
| Mean single sequence MFE | -18.69 |
| Consensus MFE | -8.61 |
| Energy contribution | -8.83 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.90 |
| Mean z-score | -0.69 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.783895 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
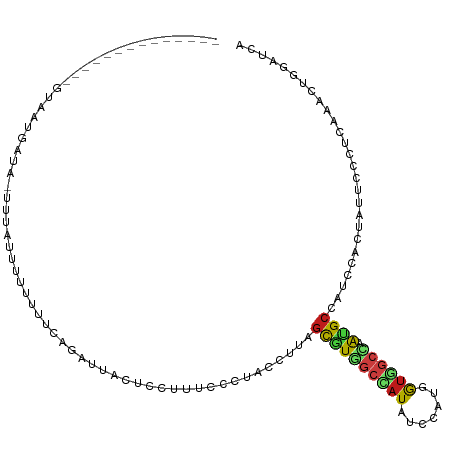
>dm3.chr3R 27601339 100 - 27905053 --------------GUCUUGAGA-UCUUUUCUUUUUCAGAUUACUCCUUUCCGUACCUUAGCGUGGCCAUAUCCAUGGUAGCCAAUGCCAUCCACUAUUCCCUCAAACUGGAUCA --------------((((.((((-.........))))))))........(((((......))((((........((((((.....))))))))))..............)))... ( -14.90, z-score = 0.02, R) >droSim1.chr3R 27219064 100 - 27517382 --------------GUUUUGAGA-UUUUUUUCUUUUCAGACUACUCCUUUCCCUACCUUAGCGUGGCCAUAUCCAUGGUGGCCAAUGCCAUCCACUAUUCCCUCAAACUGGAUCA --------------((((.((((-......))))...))))...................((((((((((.......)))))).)))).(((((..............))))).. ( -19.24, z-score = -1.33, R) >droSec1.super_31 255824 101 - 568655 --------------GUUUUGAGAUUUUUUUUCUUUUCAGACUACUCCUUUCCGUACCUUAGCGUGGCCAUAUCCAUGGUGGCCAAUGCCAUCCACUAUUCCCUCAAACUGGAUCA --------------..(((((((..........)))))))....................((((((((((.......)))))).)))).(((((..............))))).. ( -18.94, z-score = -0.90, R) >droYak2.chr3R 28535868 100 - 28832112 --------------GUGUUGACA-UGUAUACCUUUUCAGAUUACUCUUUUCCCUACCUUAGCGUGGCCAUAUCCAUGGUGGCCAAUGCCAUCCACUAUUCCCUCAAACUGGAUCA --------------((((.....-...)))).............................((((((((((.......)))))).)))).(((((..............))))).. ( -16.44, z-score = 0.41, R) >droEre2.scaffold_4820 10100610 100 + 10470090 --------------GCAUUGACA-UAUUUACCUUUUCAGAUUACUCUUUUCCCUACCUUAGCGUGGCCAUAUCCAUGGUGGCCAAUGCCAUCCACUAUUCUCUCAAACUGGAUCA --------------((((((...-.............(((....)))...............((.(((((....))))).)))))))).(((((..............))))).. ( -16.14, z-score = -0.43, R) >droAna3.scaffold_12911 1879685 115 + 5364042 AUGUCUCACUUGAAGAAAUCUGGUGGUAAUAUUUUCCAGAUUACACCUUUCCAUACCUAAGCGUGGCCAUUUCUAUGGUGGCCAAUGCCAUUCACUAUUCCCUUAAGCUUGACCA ..(((...(((((.(.(((((((..(......)..))))))).)................(((((((((((.....))))))).))))..............)))))...))).. ( -28.20, z-score = -1.88, R) >dp4.chr2 14046111 99 - 30794189 ----------------GACUAAAUAUCAUUGUCUUACAGACUACUCCUUUCCGUAUCUUAGCAUGGCCAUUUCUAUGGUGGCCAAUGCCAUACACUAUUCCCUCAAACUGGACCA ----------------..............(((.....)))...........((((....(((((((((((.....))))))).)))).)))).....(((........)))... ( -19.70, z-score = -1.72, R) >droPer1.super_0 5427230 99 + 11822988 ----------------GACUAAAUAUCAUUGUCUUACAGACUACUCCUUUCCGUAUCUUAGCGUGGCCAUUUCUAUGGUGGCCAAUGCCAUACACUAUUCCCUCAAACUGGACCA ----------------..............(((.....)))...........((((....(((((((((((.....))))))).)))).)))).....(((........)))... ( -19.30, z-score = -1.33, R) >droWil1.scaffold_181130 7502401 105 + 16660200 ----------ACUUUAUAUCAUUUACUGUGAUCUUUCAGAUUAUUCGUUUCCCUAUCUCAGUGUGGCCAUAUCGAUGGUCGCCAACGCCAUUCACUAUUCACUGAAAAUAGAUCA ----------.................((((((.....))))))........((((.((((((.((((((....))))))((....))...........))))))..)))).... ( -19.20, z-score = -1.23, R) >droVir3.scaffold_13047 11133916 106 - 19223366 ---------AGCUCAUAAUAUUGACUUAAUUUUUUUUAGAUUAUUCGUUUCCUUAUCUAAGUAUCGCUAUGUCAAUGGUGGCCAACGCCAUUCACUACUCUUUAAAGCUAGAUCA ---------((((.....(((((((.........(((((((.............))))))).........)))))))(((((....)))))..............))))...... ( -18.89, z-score = -1.75, R) >droMoj3.scaffold_6540 3523059 106 + 34148556 ---------AAAUUGUACUCAUAGCCGUCUUUCUUGCAGAUUAUUCGUUUCCCUAUUUAAGCAUAGCUAUGUCCAUGGUAGCCAACGCUAUACACUACUCCUUAAAGCUGGAUCA ---------....(((...((((((.((((.......)))).....((((........))))...))))))......(((((....))))))))....(((........)))... ( -14.20, z-score = 1.05, R) >droGri2.scaffold_14624 1026389 87 - 4233967 ----------------------------UUUUGUUGUAGAUUAUUCAUUCCCAUAUUUGAGCAUUGCCAUGUCUAUGGUGGCCAAUGCAAUACACUACUCCCUGAAGCUGGAUCA ----------------------------.......((((.....(((..........)))((((((((((.......)))).))))))......))))(((........)))... ( -16.80, z-score = 0.37, R) >anoGam1.chr2R 5293185 114 - 62725911 -GUUGUGACAUCAACGAAUCAUCGGGUCUCUUUCUACAGACUACACCUUCCCCUACUUGAGCAUCCUCAUUUCGGUGGUGUCGAAUGCAUUGCACUUUUCGAUCAAGCUCGAUCA -((..((((((((.((((......(((((........)))))...............((((....)))).)))).))))))))...))............((((......)))). ( -25.60, z-score = -0.71, R) >triCas2.ChLG7 12801469 100 + 17478683 ---------------CACUUUUGAUUGGUUUUGUUCCAGAUUAUGCGUUCCCAAUUUUGGUCGUUAUUUUGUCAGUUAUUUCGUGUGCUUAUCACUUUGCCCUCAAAAUCAACCA ---------------......((.(((((((((...((((....(((...(((....))).)))...))))...........(((.......)))........))))))))).)) ( -14.10, z-score = -0.18, R) >consensus ______________GUAAUGAUA_UUUAUUUUUUUUCAGAUUACUCCUUUCCCUACCUUAGCGUGGCCAUAUCCAUGGUGGCCAAUGCCAUCCACUAUUCCCUCAAACUGGAUCA ............................................................((((((((((.......)))))).))))........................... ( -8.61 = -8.83 + 0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:00:39 2011