
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 27,588,277 – 27,588,431 |
| Length | 154 |
| Max. P | 0.952072 |

| Location | 27,588,277 – 27,588,431 |
|---|---|
| Length | 154 |
| Sequences | 5 |
| Columns | 159 |
| Reading direction | forward |
| Mean pairwise identity | 74.60 |
| Shannon entropy | 0.45121 |
| G+C content | 0.55927 |
| Mean single sequence MFE | -47.69 |
| Consensus MFE | -30.40 |
| Energy contribution | -31.84 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.952072 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
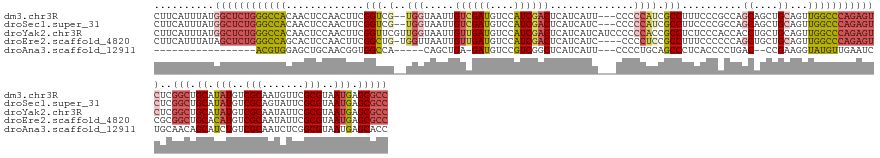
>dm3.chr3R 27588277 154 + 27905053 CUUCAUUUAUGGCUCUGGGCCACAACUCCAACUUCGGUCG--UGGUAAUUGUCGAUGUCCAUCGACUCAUCAUU---CCCCCAUCGCCUUUCCCGCCAGCAGCUGCAGUUGGCCCAGAGUCUCGGCUGCAUAUGUCGCAAUGUUCGCGUAAUGAGCGCC ..........((((((((((((..(((.((.((.(((..(--((((....((((((....))))))..))))).---.)).....((.......))..).)).)).)))))))))))))))..(((.((.(((..(((.......)))..))).))))) ( -50.00, z-score = -2.09, R) >droSec1.super_31 242826 154 + 568655 CUUCAUUUAUGGCUCUGGGCCACAACUCCAACUUCGGUCG--UGGUAAUUGUCGAUGUCCAUCGACUCAUCAUC---CCCCCAUCGCCUUCCCCGCCAGCAGCUGCAGUUGGCCCAGAGUCUCGGCUGCAUAUGUCGCAGUAUUCGCGUAAUGAGCGCC ..........((((((((((((..(((.((.((.(((..(--((((....((((((....))))))..))))).---.)).....((.......))..).)).)).)))))))))))))))...(((((.......)))))....((((.....)))). ( -51.10, z-score = -2.43, R) >droYak2.chr3R 28522721 159 + 28832112 CUUCAUUUAUGGCUCUGGGCCACAACUCCAACUUCGGUUCGUUGGUAAUUGUUGAUGUCCAUCGACUCAUCAUCAUCCCCCCACCGCCUCUCCCACCACCUGCUGCAGUUGGCCCAGAGUCUCGGCUGCAUAUGUCGCAAUAUUCGCGUAAUGAGCGCC ..........((((((((((((..(((.((((....)))((.(((....((.(((((..........))))).)).....))).))..................).)))))))))))))))..(((.((.(((..(((.......)))..))).))))) ( -45.60, z-score = -1.83, R) >droEre2.scaffold_4820 10087827 154 - 10470090 CUUCAUUUAUAGCUCUGGGCCAGCACUCCAACUUCGGCUG-UGGUUAAUUGUUGAUGUCCAUCGACUCAUCAUC----CCCCUCCGCCUUUCCCCCCAGCUGCUGCAGUUGGCCCAGAGUCGCGGCUGCACAUGUCGCAAUAUUCGCGUAAUGAGCGCC ...........(((((((((((((...........(((.(-.((.....((.(((.(((....)))))).))..----..)).).)))..........((....)).))))))))))))).(((((.......))))).......((((.....)))). ( -53.10, z-score = -3.15, R) >droAna3.scaffold_12911 1867455 131 - 5364042 -----------------ACGUGGAGCUGCAACGGUGGCCA-----CAGCUGA-GAUGUCCGUCGGCUCAUCAUU---CCCCUGCAGCCCUCACCCCUGAC--CCGAAGGUAUGUUGAAUCUGCAACAGCAUCUGUCGCAAUCUCGGCGUAAUGAGCACC -----------------..(((..((((((..((......-----.((((((-........)))))).......---))..)))))).((((..(((...--....)))((((((((...(((.((((...)))).)))...)))))))).))))))). ( -38.64, z-score = 0.53, R) >consensus CUUCAUUUAUGGCUCUGGGCCACAACUCCAACUUCGGUCG__UGGUAAUUGUCGAUGUCCAUCGACUCAUCAUC___CCCCCACCGCCUUUCCCCCCAGCAGCUGCAGUUGGCCCAGAGUCUCGGCUGCAUAUGUCGCAAUAUUCGCGUAAUGAGCGCC ..........((((((((((((........(((.((((............((((((....))))))...................((...........)).)))).)))))))))))))))..(((.((.(((..(((.......)))..))).))))) (-30.40 = -31.84 + 1.44)
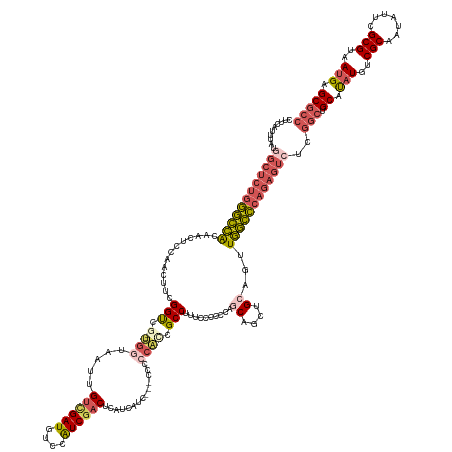
| Location | 27,588,277 – 27,588,431 |
|---|---|
| Length | 154 |
| Sequences | 5 |
| Columns | 159 |
| Reading direction | reverse |
| Mean pairwise identity | 74.60 |
| Shannon entropy | 0.45121 |
| G+C content | 0.55927 |
| Mean single sequence MFE | -54.76 |
| Consensus MFE | -33.26 |
| Energy contribution | -33.82 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.947486 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
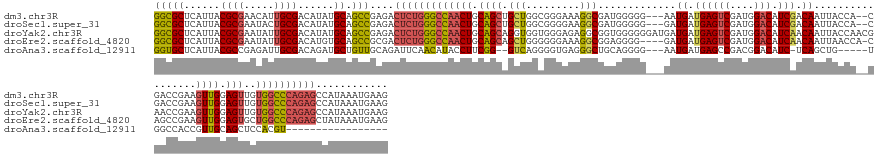
>dm3.chr3R 27588277 154 - 27905053 GGCGCUCAUUACGCGAACAUUGCGACAUAUGCAGCCGAGACUCUGGGCCAACUGCAGCUGCUGGCGGGAAAGGCGAUGGGGG---AAUGAUGAGUCGAUGGACAUCGACAAUUACCA--CGACCGAAGUUGGAGUUGUGGCCCAGAGCCAUAAAUGAAG (((((......(((((...)))))......)).)))..(.(((((((((((((.(((((.....(((......((.(((...---(((.....((((((....)))))).))).)))--)).))).))))).)))..)))))))))).).......... ( -55.30, z-score = -2.22, R) >droSec1.super_31 242826 154 - 568655 GGCGCUCAUUACGCGAAUACUGCGACAUAUGCAGCCGAGACUCUGGGCCAACUGCAGCUGCUGGCGGGGAAGGCGAUGGGGG---GAUGAUGAGUCGAUGGACAUCGACAAUUACCA--CGACCGAAGUUGGAGUUGUGGCCCAGAGCCAUAAAUGAAG (((((......((((.....))))......)).)))..(.(((((((((((((.((((((((..(...)..)))..(((..(---(.((((..((((((....)))))).)))))).--...))).))))).)))..)))))))))).).......... ( -56.50, z-score = -2.14, R) >droYak2.chr3R 28522721 159 - 28832112 GGCGCUCAUUACGCGAAUAUUGCGACAUAUGCAGCCGAGACUCUGGGCCAACUGCAGCAGGUGGUGGGAGAGGCGGUGGGGGGAUGAUGAUGAGUCGAUGGACAUCAACAAUUACCAACGAACCGAAGUUGGAGUUGUGGCCCAGAGCCAUAAAUGAAG (((((......(((((...)))))......)).)))..(.(((((((((((((.((((...((((........((.(((..(..((.((((..(((....))))))).)).)..))).)).))))..)))).)))..)))))))))).).......... ( -53.10, z-score = -1.47, R) >droEre2.scaffold_4820 10087827 154 + 10470090 GGCGCUCAUUACGCGAAUAUUGCGACAUGUGCAGCCGCGACUCUGGGCCAACUGCAGCAGCUGGGGGGAAAGGCGGAGGGG----GAUGAUGAGUCGAUGGACAUCAACAAUUAACCA-CAGCCGAAGUUGGAGUGCUGGCCCAGAGCUAUAAAUGAAG (((((.(((..(((((...)))))..))).)).)))..(.(((((((((((((.((((..((....))...(((.(.((..----(((..((((((....))).)))...)))..)).-).)))...)))).)))..)))))))))))........... ( -56.00, z-score = -1.88, R) >droAna3.scaffold_12911 1867455 131 + 5364042 GGUGCUCAUUACGCCGAGAUUGCGACAGAUGCUGUUGCAGAUUCAACAUACCUUCGG--GUCAGGGGUGAGGGCUGCAGGGG---AAUGAUGAGCCGACGGACAUC-UCAGCUG-----UGGCCACCGUUGCAGCUCCACGU----------------- .(((....((((.(((((.(((((((((...))))))))).))).....(((....)--))..)).))))(((((((((.((---.....((.(((.((((.(...-...))))-----)))))))).))))))))))))..----------------- ( -52.90, z-score = -1.61, R) >consensus GGCGCUCAUUACGCGAAUAUUGCGACAUAUGCAGCCGAGACUCUGGGCCAACUGCAGCAGCUGGCGGGAAAGGCGGUGGGGG___GAUGAUGAGUCGAUGGACAUCAACAAUUACCA__CGACCGAAGUUGGAGUUGUGGCCCAGAGCCAUAAAUGAAG (((((......((((.....))))......)).)))....(((((((((((((.((((.(((.........))).............((.((((((....))).))).)).................)))).)))..))))))))))............ (-33.26 = -33.82 + 0.56)
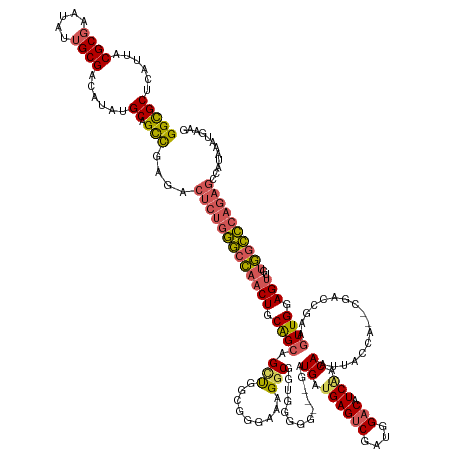
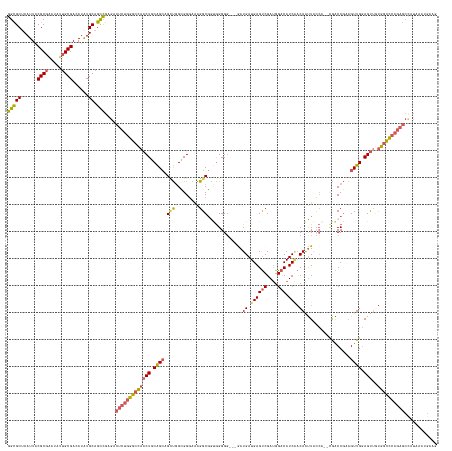
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:00:38 2011