
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 9,391,840 – 9,391,932 |
| Length | 92 |
| Max. P | 0.769163 |

| Location | 9,391,840 – 9,391,932 |
|---|---|
| Length | 92 |
| Sequences | 7 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 71.63 |
| Shannon entropy | 0.56214 |
| G+C content | 0.42510 |
| Mean single sequence MFE | -20.59 |
| Consensus MFE | -12.16 |
| Energy contribution | -12.27 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.50 |
| Mean z-score | -0.90 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.769163 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 9391840 92 + 23011544 -----AACAAGCACACAGAUACUUCCUCACAGCUUACACUGCACGAAA-CAUUGAUUGGUUUAGCAUAGAGCCAAAAGCAAAUGGAUAAAGUGAGCAA -----.....((.(((.......(((.....((((.((.((.......-)).)).(((((((......)))))))))))....)))....))).)).. ( -16.60, z-score = -0.51, R) >droSim1.chr2L 9170871 92 + 22036055 -----AACAAGCGCACAGAUACUUGCUCACAGCUUACACUGCACGAAA-CAUUGAUUGGUUUAGCAUAGAGCCAAGAGCGAAUGGAUAAAGUGAGCAG -----..................(((((((.((.......))......-(((((.(((((((......)))))))...).))))......))))))). ( -22.30, z-score = -1.35, R) >droSec1.super_3 4848653 92 + 7220098 -----AACAAGCACACAGAUACUUGCUCACAGCUUACACUGCACGAAA-CAUUGAUUGGUUUAGCAUAGAGCCAAGAGCAAAUGGAUGAAGUGAGCAG -----..................(((((((.((.......))......-(((((.(((((((......)))))))...).))))......))))))). ( -22.30, z-score = -1.56, R) >droYak2.chr2L 12065629 92 + 22324452 -----AACAAGCACACAGAUACUUCCUCACAGUUUACACUGCACGAAA-CAUUGAUUGGUUUAGCAUUGAGCCAAGAGCGAAUGGAUAAAGUGAGCAG -----.....((.(((..((..((((((.((((....)))).......-......((((((((....))))))))))).)))...))...))).)).. ( -18.70, z-score = -0.88, R) >droEre2.scaffold_4929 10003948 92 + 26641161 -----AGCCAGCACACAGAUACUUGCUCACAGCUUACACUGCACGAAA-CAUUGAUUGGCUUAGCAUAGAGCCAAAAGCAAAUGGAUAAAGUGAGCAG -----..................(((((((.((.......))......-(((((.(((((((......)))))))...).))))......))))))). ( -23.60, z-score = -1.62, R) >droAna3.scaffold_12943 744700 95 + 5039921 -AACACUCAGAUACUCAGGCACUGGCACACAGCUUACACUGAACGAAA-CAUUGAUUGGUUUUUCGCAGAGCUAAAAGCAAUCGGA-AAAGUGAGCAG -....((((.....((((.....(((.....)))....))))......-..(((((((.((((..((...)).)))).))))))).-....))))... ( -17.90, z-score = 0.12, R) >droWil1.scaffold_180772 1318718 97 - 8906247 UAUGGAGUGUAGGGGGAGGUGCCAGUAUGUCAGCAGUGUAAAACAAAAGUAUUGAUUGCUUUUUU-UGCUGUUGCAUAAAACAAAACCAAGCAAACAU .(((...(((..((....((.....((((.((((((((......(((((((.....)))))))..-))))))))))))..))....))..)))..))) ( -22.70, z-score = -0.48, R) >consensus _____AACAAGCACACAGAUACUUGCUCACAGCUUACACUGCACGAAA_CAUUGAUUGGUUUAGCAUAGAGCCAAAAGCAAAUGGAUAAAGUGAGCAG ........................((((((.((.......)).........(((.(((((((......)))))))...))).........)))))).. (-12.16 = -12.27 + 0.11)

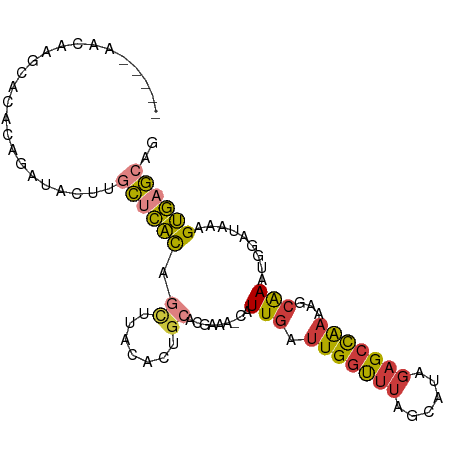
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:28:06 2011