| Sequence ID | dm3.chr3R |
|---|---|
| Location | 27,556,332 – 27,556,425 |
| Length | 93 |
| Max. P | 0.938715 |

| Location | 27,556,332 – 27,556,425 |
|---|---|
| Length | 93 |
| Sequences | 12 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 77.65 |
| Shannon entropy | 0.48658 |
| G+C content | 0.67089 |
| Mean single sequence MFE | -37.81 |
| Consensus MFE | -19.05 |
| Energy contribution | -19.10 |
| Covariance contribution | 0.05 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.938715 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
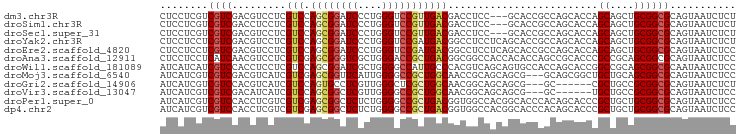
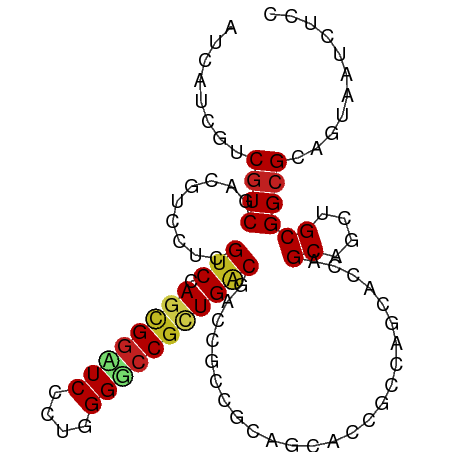
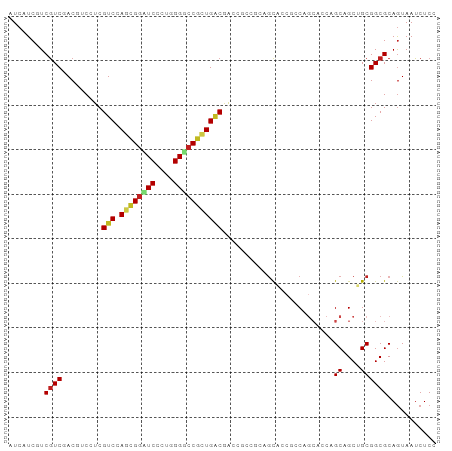
>dm3.chr3R 27556332 93 + 27905053 CUCCUCGUCGUCGACGUCCUCGUCCAGCGGAUCCCUGGGUCCGUUGACGACCUCC---GCACCGCCAGCACCAGCAGCUGCGGCGCAGUAAUCUCU ......((((((((((....)))).((((((((....))))))))))))))....---((.((((.(((.......))))))).)).......... ( -40.00, z-score = -3.67, R) >droSim1.chr3R 27173401 93 + 27517382 CUCCUCGUCGUCGACCUCCUCGUCCAGCGGAUCCCUGGGUCCGUUGACGACCUCC---GCACCGCCAGCACCAGCAGCUGCGGCGCAGUAAUCUCU ......(((((((((......))).((((((((....))))))))))))))....---((.((((.(((.......))))))).)).......... ( -37.20, z-score = -3.12, R) >droSec1.super_31 210949 93 + 568655 CUCCUCGUCGUCGACGUCCUCGUCCAGCGGAUCCCUGGGUCCGUUGACGACCUCC---GCACCGCCAGCACCAGCAGCUGCGGCGCAGUAAUCUCU ......((((((((((....)))).((((((((....))))))))))))))....---((.((((.(((.......))))))).)).......... ( -40.00, z-score = -3.67, R) >droYak2.chr3R 28490844 96 + 28832112 CUCCUCCUCGUCGACGUCCUCGUCCAGCGGAUCCCUGGGUCCGAUGACGGCCUCCUCAGCACCGCCAGCACCAGCAGCUGCGGCGCAGUAAUCUCU .......(((((((((....))))...((((((....))))))..)))))........((.((((.(((.......))))))).)).......... ( -33.50, z-score = -1.11, R) >droEre2.scaffold_4820 10050397 96 - 10470090 CUCCUCCUCGUCGACGUCCUCGUCCAGCGGAUCCCUGGGUCCGAUGACGGCCUCCUCAGCACCGCCAGCACCAGCAGCUGCGGCGCAGUAAUCUCC .......(((((((((....))))...((((((....))))))..)))))........((.((((.(((.......))))))).)).......... ( -33.50, z-score = -1.20, R) >droAna3.scaffold_12911 1828367 96 - 5364042 CUCCUCCUCAUCAACGUCCUCGUCGAGCGGGUCGCUGGGACCGCUGACGGCGGCCACCACACCAGCCGCACCCGCCGCAGCGGCGCAGUAAUCUCC ..(((.(((....(((....))).))).)))..((((.(.((((((.((((((..................))))))))))))).))))....... ( -35.77, z-score = -0.80, R) >droWil1.scaffold_181089 9523733 96 + 12369635 AUCAUCAUCGUCCACCUCCUCGUCCAGCGGAUCGCUGGGGCCAUUGCCCACGUCAGCAGUGCCACCAGCACCGGCCGCAGCGGCGCAAUAAUCUCC ....................(((((.((((...((((((((.(((((........)))))))).))))).....)))).).))))........... ( -30.50, z-score = -1.03, R) >droMoj3.scaffold_6540 22172241 93 - 34148556 AUCAUCGUCGUCGACGUCAUCGUCGAGCGGUUCAUUGGGGCCGCUGGCAACCGCAGCAGCG---GCAGCGGCUGCUGCAGCGGCGCAGUAAUCUCC ......(((.((((((....))))))(((((((....))))))).)))..((((.((((((---((....)))))))).))))............. ( -44.70, z-score = -2.77, R) >droGri2.scaffold_14906 11856116 87 - 14172833 AUCAUCGUCGUCCACGUCAUCGUCCAGUGCCUCGUUGGGCUCGCUGGCAACGGCAGCAGCG---GC------CGCUGCCGCGGCGCAGUAAUCUCU ......(.((....)).)........(((((.......(((.((((....))))))).(((---((------....)))))))))).......... ( -32.10, z-score = -0.46, R) >droVir3.scaffold_13047 9828125 87 - 19223366 AUCAUCGUCGUCGACAUCAUCGUCCAGCGGCUCGUUGGGGCCGCUGGCAACGGCAGCAGCG---GC------UGCUGCCGCGGCGCAGUAAUCUCC ......(.(((((........(.((((((((((....)))))))))))..(((((((((..---.)------)))))))))))))).......... ( -44.40, z-score = -3.53, R) >droPer1.super_0 3847471 96 + 11822988 AUCAUCGUCGUCCACCUCGUCGUCGAGCGGCUCUCUGGGGCCGCUGACGGUGGCCACGGCACCCACAGCACCCGCUGCUGCGGCGCAGUAAUCUCC .....(((.(.(((((.....(((.((((((((....))))))))))))))))).)))((.((((((((....)))).)).)).)).......... ( -41.00, z-score = -1.55, R) >dp4.chr2 14244711 96 - 30794189 AUCAUCGUCGUCCACCUCGUCGUCGAGCGGCUCUCUGGGGCCGCUGACGGUGGCCACGGCACCCACAGCACCCGCUGCUGCGGCGCAGUAAUCUCC .....(((.(.(((((.....(((.((((((((....))))))))))))))))).)))((.((((((((....)))).)).)).)).......... ( -41.00, z-score = -1.55, R) >consensus AUCAUCGUCGUCGACGUCCUCGUCCAGCGGAUCCCUGGGGCCGCUGACGACCGCCGCAGCACCGCCAGCACCAGCAGCUGCGGCGCAGUAAUCUCC ........((((.........(((.((((((((....))))))))))).........................((....))))))........... (-19.05 = -19.10 + 0.05)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:00:36 2011