
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 27,535,811 – 27,535,904 |
| Length | 93 |
| Max. P | 0.697950 |

| Location | 27,535,811 – 27,535,904 |
|---|---|
| Length | 93 |
| Sequences | 8 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 75.34 |
| Shannon entropy | 0.49216 |
| G+C content | 0.58885 |
| Mean single sequence MFE | -35.32 |
| Consensus MFE | -21.73 |
| Energy contribution | -22.27 |
| Covariance contribution | 0.55 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.05 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.697950 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 27535811 93 + 27905053 AACUUUGUCGCUUUGUGGGCGGCAUCAAAGUUGCCGUUCACUUGCAAU------UGCGAUCCGAUGGUGGGC-GGCGUGGAGCGGUUUGGCUCCCCUAGA--- ((((((((((((.....)))))))...)))))(((((((((((((...------.))).......)))))))-)))(.(((((......))))).)....--- ( -36.61, z-score = -1.29, R) >dp4.chr2 14222093 98 - 30794189 --UGUUGUCGCCUUGUGGGCGUCAUCAAAGUUGCCGUUCACUUGCGAUAUUCCUCUCGGCUUGGUGCCGGGCUGGUAGGCUGGUGGGCGGCGCCCCAGCG--- --((.((.((((.....)))).)).)).....((((((((((.((..((((...((((((.....))))))..)))).)).))))))))))((....)).--- ( -42.00, z-score = -1.13, R) >droAna3.scaffold_12911 1807906 91 - 5364042 AACUUUGUCGCCUUGUGGGCGGCAUCAAAGUUGCCGUUCACUUGCAAU------UGCGAUCCGCUGGUGGGC-GGCGUGGUGA-----GGCUUUGCUUGGACC ..((..((.(((((((((((((((.......)))))))))).....((------..((..(((((....)))-))))..))))-----)))...))..))... ( -36.10, z-score = -1.29, R) >droEre2.scaffold_4820 10029161 88 - 10470090 AACUUUGUCGCUUUGUGGGCGGCAUCAAAGUUGCCGUUCACUUGCAAU------UGCGAUCCGAUGGUGGGC-GGCGUGGCGC-----GGCUCCUCUAGA--- ......(((((...((((((((((.......)))))))))).......------.)))))....(((.((((-.(((...)))-----.)).)).)))..--- ( -32.60, z-score = -0.58, R) >droYak2.chr3R 28469098 88 + 28832112 AGCUUUGUCGCUUGGUGGGCGGCAUCAAAGUUGCCGUUCACUUGCAAU------UGCGAUCCGAUGGUGGGC-GGCGUGGAGC-----GGCUCCUCUAGA--- ......(((((..(((((((((((.......)))))))))))......------.)))))....(((.((((-.((.....))-----.)).)).)))..--- ( -34.10, z-score = -0.63, R) >droSec1.super_31 190314 93 + 568655 AACUUUGUCGCUUUGUGGGCGGCAUCAAAGUUGCCGUUCACUUGCAAU------UGCGAUCCGAUGGUGGGC-GGCGUGGAGCGGCUCGGCUCCUCUAGG--- ((((((((((((.....)))))))...)))))(((((((((((((...------.))).......)))))))-)))(.(((((......))))).)....--- ( -37.41, z-score = -1.46, R) >droSim1.chr3R 27150752 93 + 27517382 AACUUUGUCGCUUUGUGGGCGGCAUCAAAGUUGCCGUUCACUUGCAAU------UGCGAUCCGAUGGUGGGC-GGCGUGGAGCGGCUCGGCUCCUCUAGA--- ((((((((((((.....)))))))...)))))(((((((((((((...------.))).......)))))))-)))(.(((((......))))).)....--- ( -37.41, z-score = -1.41, R) >droVir3.scaffold_13047 9797995 72 - 19223366 UGCUUCGCAUCAUUGUGGGCGUCAUCAAAGUUGCCGUUCACCAGAAAU-------GUGGGCGGCUGA--GGC-UGC-UGCCGC-------------------- .(((.((((....)))))))............(((((((((.(....)-------)))))))))...--(((-...-.)))..-------------------- ( -26.30, z-score = -0.61, R) >consensus AACUUUGUCGCUUUGUGGGCGGCAUCAAAGUUGCCGUUCACUUGCAAU______UGCGAUCCGAUGGUGGGC_GGCGUGGAGC_____GGCUCCUCUAGA___ .....((((((((.((((((((((.......))))))))))(((((........))))).........)))).)))).((((........))))......... (-21.73 = -22.27 + 0.55)

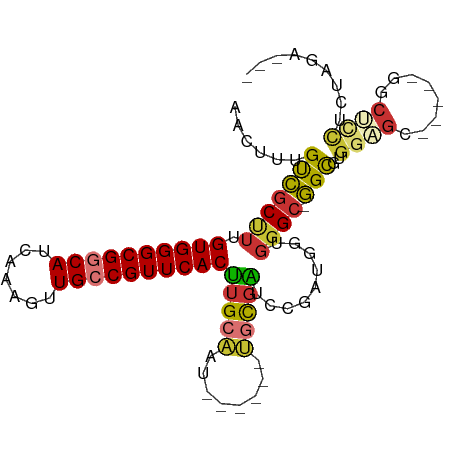
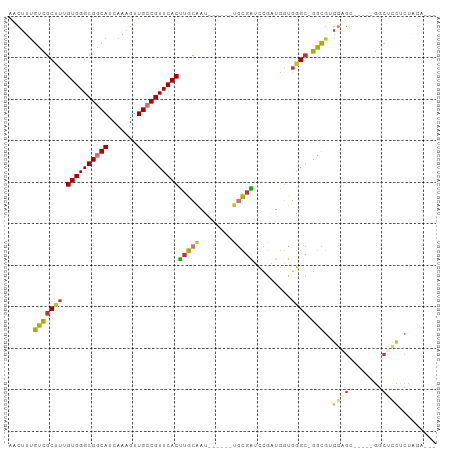
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:00:34 2011