
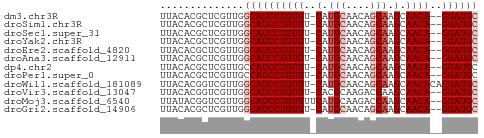
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 27,508,805 – 27,508,855 |
| Length | 50 |
| Max. P | 0.867144 |

| Location | 27,508,805 – 27,508,855 |
|---|---|
| Length | 50 |
| Sequences | 12 |
| Columns | 53 |
| Reading direction | forward |
| Mean pairwise identity | 95.74 |
| Shannon entropy | 0.09114 |
| G+C content | 0.49922 |
| Mean single sequence MFE | -17.60 |
| Consensus MFE | -9.43 |
| Energy contribution | -9.84 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.23 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.662253 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
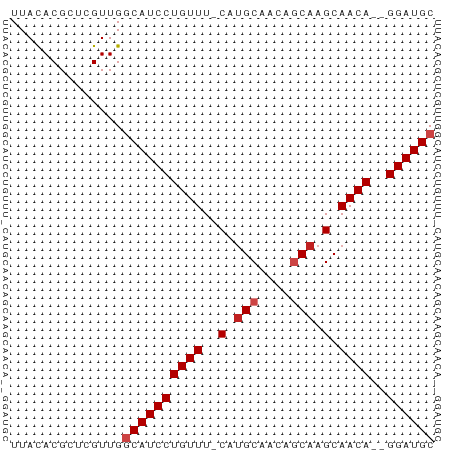
>dm3.chr3R 27508805 50 + 27905053 UUACACGCUCGUUGGCAUCCUGUUU-CAUGCAACAGCAAGCAACA--GGAUGC ..............((((((((((.-(.(((....))).).))))--)))))) ( -18.80, z-score = -3.67, R) >droSim1.chr3R 27128805 50 + 27517382 UUACACGCUCGUUGGCAUCCUGUUU-CAUGCAACAGCAAGCAACA--GGAUGC ..............((((((((((.-(.(((....))).).))))--)))))) ( -18.80, z-score = -3.67, R) >droSec1.super_31 168389 50 + 568655 UUACACGCUCGUUGGCAUCCUGUUU-CAUGCAACAGCAAGCAACA--GGAUGC ..............((((((((((.-(.(((....))).).))))--)))))) ( -18.80, z-score = -3.67, R) >droYak2.chr3R 28446346 50 + 28832112 UUACACGCUCGUUGGCAUCCUGUUU-CAUGCAACAGCAAGCAACA--GGAUGC ..............((((((((((.-(.(((....))).).))))--)))))) ( -18.80, z-score = -3.67, R) >droEre2.scaffold_4820 10004362 50 - 10470090 UUACACGCUCGUUGGCAUCCUGUUU-CAUGCAACAGCAAGCAACA--GGAUGC ..............((((((((((.-(.(((....))).).))))--)))))) ( -18.80, z-score = -3.67, R) >droAna3.scaffold_12911 1781102 50 - 5364042 UUACACGCUCGUUGGCAUCCUGUUU-CAUGCAACAGCAAGCAACA--GGAUGC ..............((((((((((.-(.(((....))).).))))--)))))) ( -18.80, z-score = -3.67, R) >dp4.chr2 14189761 50 - 30794189 UUACACGCUCGUUGCCAUCCUGUUU-CAUGCAACAGCAAGCAACA--GGAUGC ......((.....))(((((((((.-(.(((....))).).))))--))))). ( -17.00, z-score = -3.24, R) >droPer1.super_0 3791377 50 + 11822988 UUACACGCUCGUUGCCAUCCUGUUU-CAUGCAACAGCAAGCAACA--GGAUGC ......((.....))(((((((((.-(.(((....))).).))))--))))). ( -17.00, z-score = -3.24, R) >droWil1.scaffold_181089 9472634 52 + 12369635 UUACACGGUCGUUGGCAUCCUGUUU-CAUGCAACAGCAAGCAACACAGGAUGC ..............(((((((((..-..(((........)))..))))))))) ( -15.70, z-score = -2.27, R) >droVir3.scaffold_13047 9760783 50 - 19223366 UUACACGGUCGUUGGCAUCCUGUUU-CACGCAAGACCAAGCAACA--GGAUGC ..............((((((((((.-...((........))))))--)))))) ( -14.80, z-score = -2.08, R) >droMoj3.scaffold_6540 22114147 51 - 34148556 UUAUACGGUCGUUGGCAUCCUGUUUUCAUGCAAGACCAAGCAACA--GGAUGC ..............(((((((((.....(((........))))))--)))))) ( -15.10, z-score = -2.27, R) >droGri2.scaffold_14906 11798653 50 - 14172833 UUACACGCUCGUUGGCAUCCUGUUU-CAUGCAACAGCAAGCAACA--GGAUGC ..............((((((((((.-(.(((....))).).))))--)))))) ( -18.80, z-score = -3.67, R) >consensus UUACACGCUCGUUGGCAUCCUGUUU_CAUGCAACAGCAAGCAACA__GGAUGC ..............((((((((((....(((....)))...))))..)))))) ( -9.43 = -9.84 + 0.42)

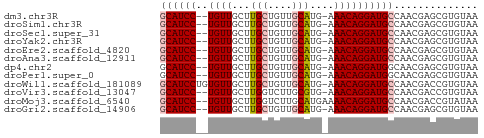
| Location | 27,508,805 – 27,508,855 |
|---|---|
| Length | 50 |
| Sequences | 12 |
| Columns | 53 |
| Reading direction | reverse |
| Mean pairwise identity | 95.74 |
| Shannon entropy | 0.09114 |
| G+C content | 0.49922 |
| Mean single sequence MFE | -19.52 |
| Consensus MFE | -12.60 |
| Energy contribution | -12.86 |
| Covariance contribution | 0.26 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.14 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.867144 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 27508805 50 - 27905053 GCAUCC--UGUUGCUUGCUGUUGCAUG-AAACAGGAUGCCAACGAGCGUGUAA ((((((--((((...(((....)))..-.)))))))))).............. ( -20.70, z-score = -3.56, R) >droSim1.chr3R 27128805 50 - 27517382 GCAUCC--UGUUGCUUGCUGUUGCAUG-AAACAGGAUGCCAACGAGCGUGUAA ((((((--((((...(((....)))..-.)))))))))).............. ( -20.70, z-score = -3.56, R) >droSec1.super_31 168389 50 - 568655 GCAUCC--UGUUGCUUGCUGUUGCAUG-AAACAGGAUGCCAACGAGCGUGUAA ((((((--((((...(((....)))..-.)))))))))).............. ( -20.70, z-score = -3.56, R) >droYak2.chr3R 28446346 50 - 28832112 GCAUCC--UGUUGCUUGCUGUUGCAUG-AAACAGGAUGCCAACGAGCGUGUAA ((((((--((((...(((....)))..-.)))))))))).............. ( -20.70, z-score = -3.56, R) >droEre2.scaffold_4820 10004362 50 + 10470090 GCAUCC--UGUUGCUUGCUGUUGCAUG-AAACAGGAUGCCAACGAGCGUGUAA ((((((--((((...(((....)))..-.)))))))))).............. ( -20.70, z-score = -3.56, R) >droAna3.scaffold_12911 1781102 50 + 5364042 GCAUCC--UGUUGCUUGCUGUUGCAUG-AAACAGGAUGCCAACGAGCGUGUAA ((((((--((((...(((....)))..-.)))))))))).............. ( -20.70, z-score = -3.56, R) >dp4.chr2 14189761 50 + 30794189 GCAUCC--UGUUGCUUGCUGUUGCAUG-AAACAGGAUGGCAACGAGCGUGUAA .(((((--((((...(((....)))..-.)))))))))((.....))...... ( -19.00, z-score = -2.46, R) >droPer1.super_0 3791377 50 - 11822988 GCAUCC--UGUUGCUUGCUGUUGCAUG-AAACAGGAUGGCAACGAGCGUGUAA .(((((--((((...(((....)))..-.)))))))))((.....))...... ( -19.00, z-score = -2.46, R) >droWil1.scaffold_181089 9472634 52 - 12369635 GCAUCCUGUGUUGCUUGCUGUUGCAUG-AAACAGGAUGCCAACGACCGUGUAA (((((((((..(((........)))..-..))))))))).............. ( -18.80, z-score = -3.03, R) >droVir3.scaffold_13047 9760783 50 + 19223366 GCAUCC--UGUUGCUUGGUCUUGCGUG-AAACAGGAUGCCAACGACCGUGUAA ((((((--((((...((......))..-.)))))))))).............. ( -16.20, z-score = -2.00, R) >droMoj3.scaffold_6540 22114147 51 + 34148556 GCAUCC--UGUUGCUUGGUCUUGCAUGAAAACAGGAUGCCAACGACCGUAUAA ((((((--((((((........))).....))))))))).............. ( -16.40, z-score = -2.82, R) >droGri2.scaffold_14906 11798653 50 + 14172833 GCAUCC--UGUUGCUUGCUGUUGCAUG-AAACAGGAUGCCAACGAGCGUGUAA ((((((--((((...(((....)))..-.)))))))))).............. ( -20.70, z-score = -3.56, R) >consensus GCAUCC__UGUUGCUUGCUGUUGCAUG_AAACAGGAUGCCAACGAGCGUGUAA ((((((..((((...(((....)))....)))))))))).............. (-12.60 = -12.86 + 0.26)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:00:29 2011