
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 27,507,750 – 27,507,853 |
| Length | 103 |
| Max. P | 0.713953 |

| Location | 27,507,750 – 27,507,853 |
|---|---|
| Length | 103 |
| Sequences | 9 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 73.23 |
| Shannon entropy | 0.54345 |
| G+C content | 0.55094 |
| Mean single sequence MFE | -34.33 |
| Consensus MFE | -14.90 |
| Energy contribution | -15.68 |
| Covariance contribution | 0.77 |
| Combinations/Pair | 1.65 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.713953 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
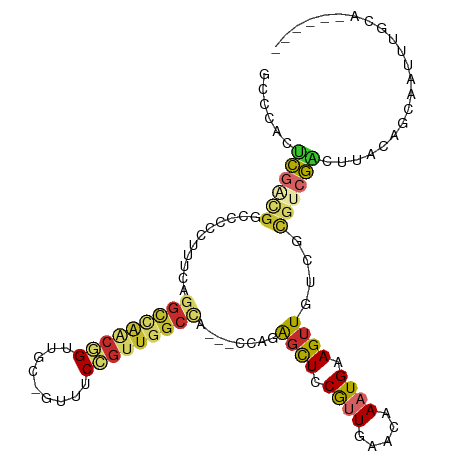
>dm3.chr3R 27507750 103 + 27905053 GCCCACUCGACGGCCCCCUUUCUGGCCAACGGUUGC-GUUUCCGUUGCCCA---CCAGAGCUCCGUUGAACAAAUGAAGUUGUCGCGGCGGCUUACAGCAAUUUGCA------ (((..(.(((((((.....((((((.((((((....-....))))))....---))))))...((((.....))))..))))))).)..))).....((.....)).------ ( -31.60, z-score = -0.40, R) >droSim1.chr3R 27127768 103 + 27517382 GCCCACUCGACGGCCCCCUUUCUGGCCAACGGUUGC-GUUUCCGUUGGCCA---CCAGAGCUCCGUUGAACAAAUGAAGUUGUCGCGGCGACUUACAGCAAUUUGCA------ ((....(((((((..(.((...((((((((((....-....))))))))))---..)).)..))))))).....(((((((((....))))))).))))........------ ( -38.70, z-score = -2.75, R) >droSec1.super_31 167359 103 + 568655 GCCCACUCGACGGCCCCCUUUCUGGCCAACGGUUGC-GUUUCCGUUGGCCA---CCAGAGCUCCGUUGAACAAAUGAAGUUGUCGCGGCGACUUACAGCAAUUUGCA------ ((....(((((((..(.((...((((((((((....-....))))))))))---..)).)..))))))).....(((((((((....))))))).))))........------ ( -38.70, z-score = -2.75, R) >droYak2.chr3R 28445272 109 + 28832112 GUCCACUCGACGGCCCCCUUUUAGGCCAACGGUUGC-AUUUCCGUUGGCCA---CCAGAGCUCCGUUGAACAAAUGAAGUUGUCGCGUCGACUUACAGCGAUUUGCAUGUGCA (((.((.(((((((.........(((((((((....-....))))))))).---.........((((.....))))..))))))).)).))).((((((.....)).)))).. ( -37.00, z-score = -2.32, R) >droEre2.scaffold_4820 10003275 103 - 10470090 ACCCACUCGACGGCCCCCUUUCAGGCCAACGGUUGC-GUUUCCGUUGGCCA---CCAGAGCUCCGCUGAGCAAAUGAAGUUGUCGCGUCGGCUUACAGCAAUUUGCA------ .((.((.(((((((.....(((((((((((((....-....))))))))).---.....((((....))))...))))))))))).)).))......((.....)).------ ( -38.80, z-score = -2.52, R) >droAna3.scaffold_12911 1779005 98 - 5364042 ---AACCCAA---UCUCUGUCUCGUCGAACGGUUGCCGUUUCCGUUGGCUG---CCACAGCUCCGUUGAACAAAUGAAGUUGUCGCGUCGACUCACAACAAUUUGCA------ ---.......---....(((...(((((..(((.((((.......)))).)---))((((((.((((.....)))).))))))....)))))..)))..........------ ( -24.80, z-score = -0.56, R) >dp4.chr2 14187798 105 - 30794189 GUUGCCUCGGUG--CCCGUUGCCCGUUGGCCGAUGCCGUUUCCGUUGGCCGACGCCGGAGCUCCGUUGAACAAAUGAAGUUGUCGCGUCGGUUUACAACAAUUUGCA------ ((((((.(((.(--(.....)))))..))).............((..((((((((.(.((((.((((.....)))).)))).).))))))))..)))))........------ ( -35.60, z-score = -0.52, R) >droPer1.super_0 3789617 105 + 11822988 GUUGCCUCGGUG--CCCGUUGCCCGUUGCCCGAUGCCGUUUCCGUUGGCCGACGCCGGAGCUCCGUUCAACAAAUGAAGUUGUCGCGUCGGUUUACAACAAUUUGCA------ (((((.((((.(--(.((.....))..)))))).)).......((..((((((((.(.((((.((((.....)))).)))).).))))))))..)))))........------ ( -34.00, z-score = -0.74, R) >droWil1.scaffold_181089 9470735 99 + 12369635 AUCAGCGUUG-GCCAUGACAACAGGCCAACAGUCGC-ACAUCUAC-AGCUA-----UUGCUUUCAAUGGCUAAAUGAAGUUGUCUUGUCACUUGCAAGUAAUUUGCA------ ......((((-(((.((....)))))))))....((-........-(((((-----(((....)))))))).....((((((.(((((.....))))))))))))).------ ( -29.80, z-score = -1.93, R) >consensus GCCCACUCGACGGCCCCCUUUCAGGCCAACGGUUGC_GUUUCCGUUGGCCA___CCAGAGCUCCGUUGAACAAAUGAAGUUGUCGCGUCGACUUACAGCAAUUUGCA______ ......((((((...........(((((((((.........)))))))))........((((.((((.....)))).))))....))))))...................... (-14.90 = -15.68 + 0.77)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:00:27 2011