
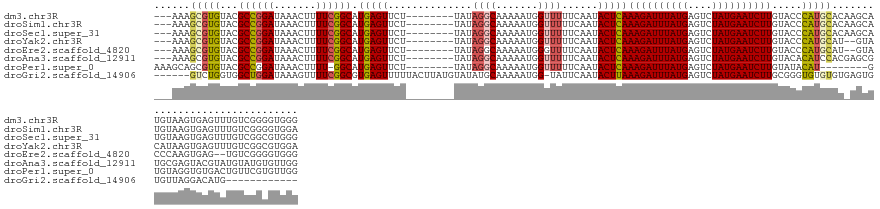
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 27,452,223 – 27,452,356 |
| Length | 133 |
| Max. P | 0.760211 |

| Location | 27,452,223 – 27,452,356 |
|---|---|
| Length | 133 |
| Sequences | 8 |
| Columns | 144 |
| Reading direction | reverse |
| Mean pairwise identity | 81.98 |
| Shannon entropy | 0.36992 |
| G+C content | 0.39733 |
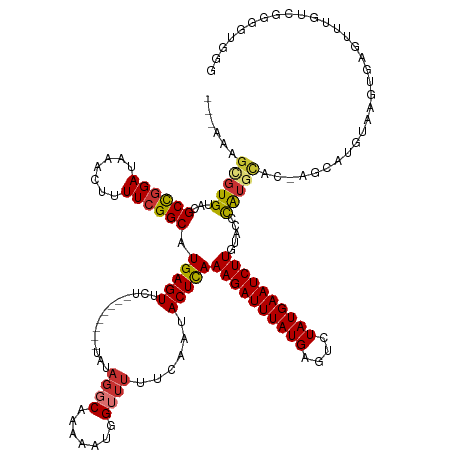
| Mean single sequence MFE | -35.86 |
| Consensus MFE | -20.86 |
| Energy contribution | -20.88 |
| Covariance contribution | 0.02 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.760211 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 27452223 133 - 27905053 ---AAAGCGUGUACGCCGGAUAAACUUUUCGGCAUGAGUUCU--------UAUAGGCAAAAAUGGUUUUUCAAUACUCAAAGAUUUAUGAGUCUAUGAAUCUUGUACCCAUGCACAAGCAUGUAAGUGAGUUUGUCGGGGUGGG ---.......((((((((((.......)))))).(((((...--------...((((.......))))......)))))((((((((((....))))))))))))))((((.(((((((..........))))))..).)))). ( -34.70, z-score = -1.20, R) >droSim1.chr3R 27072083 133 - 27517382 ---AAAGCGUGUACGCCGGAUAAACUUUUCGGCAUGAGUUCU--------UAUAGGCAAAAAUGGUUUUUCAAUACUCAAAGAUUUAUGAGUCUAUGAAUCUUGUACCCAUGCACAAGCAUGUAAGUGAGUUUGUCGGGGUGGA ---........(((.((.(((((((((..(((..(((((...--------...((((.......))))......)))))((((((((((....))))))))))...))(((((....)))))...).))))))))).))))).. ( -34.60, z-score = -1.16, R) >droSec1.super_31 112561 133 - 568655 ---AAAGCGUGUACGCCGGAUAAACUUUUCGGCAUGAGUUCU--------UAUAGGCAAAAAUGGUUUUUCAAUACUCAAAGAUUUAUGAGUCUAUGAAUCUUGUACCCAUGCACAAGCAUGUAAGUGAGUUUGUCGGCGUGGG ---.......((((((((((.......)))))).(((((...--------...((((.......))))......)))))((((((((((....))))))))))))))((((((((((((..........))))))..)))))). ( -38.50, z-score = -2.32, R) >droYak2.chr3R 28388562 131 - 28832112 ---AAAGCGUGUACGCCGGAUAAACUUUUCGGCAUGAGUUCU--------UAUAGGCAAAAAUGGUUUUUCAAUACUCAAAGAUUUAUGAGUCUAUGAAUCUUGUACCCAUGCAU--GUACAUAAGUGAGUUUGUCGGCGUGGA ---.......((((((((((.......)))))).(((((...--------...((((.......))))......)))))((((((((((....))))))))))))))((((((.(--(.(((..........))))))))))). ( -35.50, z-score = -1.81, R) >droEre2.scaffold_4820 9947762 129 + 10470090 ---AAAGCGUGUACGCCGGAUAAACUUUUCGGCAUGAGUUCU--------UAUAGGCAAAAAUGGGUUUUCAAUACUCAAAGAUUUAUGAGUCUAUGAAUCUUGUACCCAUGCAU--GUACCCAAGUGAG--UGUCGGGGUGGG ---...(.((((..((((((.......)))))).((((..((--------(((........)))))..)))))))).).((((((((((....)))))))))).(((((..((((--.(((....))).)--)))..))))).. ( -35.50, z-score = -1.24, R) >droAna3.scaffold_12911 1708164 133 + 5364042 ---AAAGCGUGUACGCCGGAUAAACUUUUCGGCAUGAGUUCU--------UAUAGGCAAAAAUGGUUUUUCAAUACUCAAAGAUUUAUGAGUCUAUGAAUCUUGUACACAUCCACGAGCGUGCGAGUACGUAUGUAUGUGUUGG ---.....((((((((((((.......)))))).(((((...--------...((((.......))))......)))))((((((((((....))))))))))))))))...((((.(((((((....))))))).)))).... ( -42.40, z-score = -3.53, R) >droPer1.super_0 3730829 127 - 11822988 AAAGCAGCGUGUACGCCGGAUAAACUUUU-GGCAUGAGUUCU--------UAUAGGCAAAAAUGGUUUUUCAAUACUCAAAGAUUUAUGAGUCUAUGAAUCUUGUAUACAU--------GUGUAGGUGUGACUGUUCGUGUUGG ....((((((((((((((((......)))-))).(((((...--------...((((.......))))......)))))((((((((((....))))))))))))))))((--------(..(((......)))..))))))). ( -34.10, z-score = -1.89, R) >droGri2.scaffold_14906 11739410 125 + 14172833 ------GUCUGGUGGCUGGAUAAAGUUUUCGGCGUGAGUUUUUACUUAUGUAUAUGCAAAAAUGG-UAUUCAAUACUUAAAGAUUUAUGAGUCUAUGAAUCUUGCGGGUGUGUGUGAGUGUGUUAGGACAUG------------ ------((((....((((((.......)))))).........((((((..(((((.(......((-(((...)))))..((((((((((....))))))))))..).)))))..)))))).....))))...------------ ( -31.60, z-score = -2.07, R) >consensus ___AAAGCGUGUACGCCGGAUAAACUUUUCGGCAUGAGUUCU________UAUAGGCAAAAAUGGUUUUUCAAUACUCAAAGAUUUAUGAGUCUAUGAAUCUUGUACCCAUGCAC_AGCAUGUAAGUGAGUUUGUCGGGGUGGG ......(((((...((((((.......)))))).(((((...........(((........)))..........)))))((((((((((....)))))))))).....)))))............................... (-20.86 = -20.88 + 0.02)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:00:20 2011