| Sequence ID | dm3.chr3R |
|---|---|
| Location | 27,443,945 – 27,444,043 |
| Length | 98 |
| Max. P | 0.996539 |

| Location | 27,443,945 – 27,444,043 |
|---|---|
| Length | 98 |
| Sequences | 7 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 70.19 |
| Shannon entropy | 0.54336 |
| G+C content | 0.44805 |
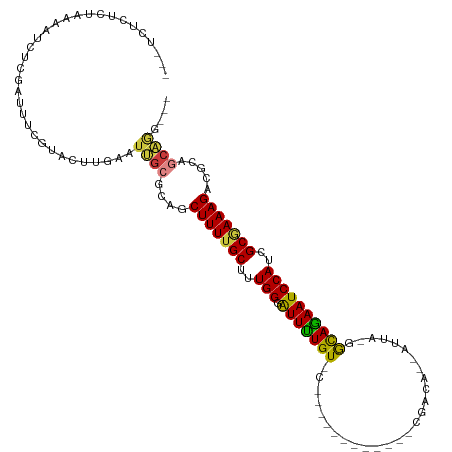
| Mean single sequence MFE | -30.47 |
| Consensus MFE | -17.51 |
| Energy contribution | -17.84 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.95 |
| SVM RNA-class probability | 0.996539 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
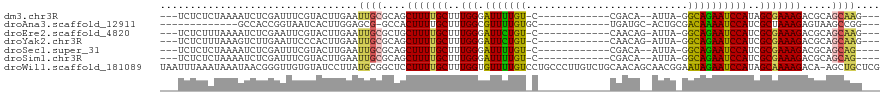
>dm3.chr3R 27443945 98 + 27905053 ---UCUCUCUAAAAUCUCGAUUUCGUACUUGAAUUGCGCAGCUUUUGCUUUGGGAUUUUGU-C------------CGACA--AUUA-GGCAGAAUCCAUAGCGAAAGACGCAGCAAG--- ---........................((((...((((...((((((((...((((((((.-(------------(....--....-))))))))))..)))))))).)))).))))--- ( -31.70, z-score = -3.18, R) >droAna3.scaffold_12911 1699492 90 - 5364042 -------------GCCACCGGUAAUCACUUGGAGCG-GCCACUUUUGCUUUGGCGUUUUGUGC------------UGAUGC-ACUGCGACAAAAUCCAUCGCUAAAGAGUAAGCCGG--- -------------....(((((....((((..((((-((((.........))))(((..((((------------....))-))...))).........))))...))))..)))))--- ( -28.00, z-score = -1.03, R) >droEre2.scaffold_4820 9939328 99 - 10470090 ---UCUCUUUAAAAUCUCGAAUUCGUACUUGAAUUGCGCUGCUUUUGCUUUGGGAUUCUGU-C------------CAACAG-AUUA-GGCAGAAUCCAUCGCGAAAGACGCAGCAAG--- ---................((((((....))))))..(((((((((((....(((((((((-(------------......-....-))))))))))...))))))...)))))...--- ( -31.20, z-score = -2.86, R) >droYak2.chr3R 28380058 99 + 28832112 ---UCUCUUUAAAGUCUUGAAUUCCCACUUGAAUUGCGCAGCUUUUGCUUUGGGAUUCUGU-C------------CAACAG-AUUA-GGCAGAAUCCAUCGCGAAAGACGCAGCAAG--- ---........................((((...((((...(((((((....(((((((((-(------------......-....-))))))))))...))))))).)))).))))--- ( -30.80, z-score = -2.60, R) >droSec1.super_31 104266 97 + 568655 ---UCUCUCUAAAAUCUCGAUUUCGUACUUGAAUUGCGCAGCUUUUGCUUUGGGAUUUUGU-C------------CGACA--AUUA-GGCAGAAUCCAUCGCGAAAGACGCAGCAG---- ---..................((((....)))).(((((..(((((((....((((((((.-(------------(....--....-))))))))))...)))))))..)).))).---- ( -27.60, z-score = -1.72, R) >droSim1.chr3R 27063823 97 + 27517382 ---UCUCUCUAAAAUCUCGAUUUCGUACUUGAAUUGCGCAGCUUUUGCUUUGGGAUUUUGU-C------------CGACA--AUUA-GGCAGAAUCCAUCGCGAAAGACGCAGCAG---- ---..................((((....)))).(((((..(((((((....((((((((.-(------------(....--....-))))))))))...)))))))..)).))).---- ( -27.60, z-score = -1.72, R) >droWil1.scaffold_181089 9399996 119 + 12369635 UAAUUUAAAUAAAUAACGGGUUGUGUAUCCUUAUGCGGCUCCUUUUGCUUUGGUGUUUUGUCCUGCCCUUGUCUGCAACAGCAACGGAAUAGAAUCCAUAGCAAAAGACA-AGCUGCUCG ............((((.((((.....))))))))((((((.((((((((.(((.((((((((((((..(((....)))..)))..)).)))))))))).))))))))...-))))))... ( -36.40, z-score = -3.29, R) >consensus ___UCUCUCUAAAAUCUCGAUUUCGUACUUGAAUUGCGCAGCUUUUGCUUUGGGAUUUUGU_C____________CGACA__AUUA_GGCAGAAUCCAUCGCGAAAGACGCAGCAGG___ .................................(((((...(((((((..(((.(((((((...........................))))))))))..))))))).)))))....... (-17.51 = -17.84 + 0.33)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:00:17 2011