
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 9,369,884 – 9,369,978 |
| Length | 94 |
| Max. P | 0.998076 |

| Location | 9,369,884 – 9,369,978 |
|---|---|
| Length | 94 |
| Sequences | 9 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 75.30 |
| Shannon entropy | 0.51827 |
| G+C content | 0.44700 |
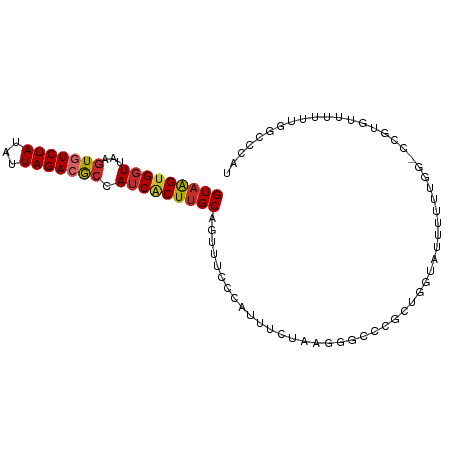
| Mean single sequence MFE | -26.54 |
| Consensus MFE | -15.18 |
| Energy contribution | -15.50 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.25 |
| SVM RNA-class probability | 0.998076 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
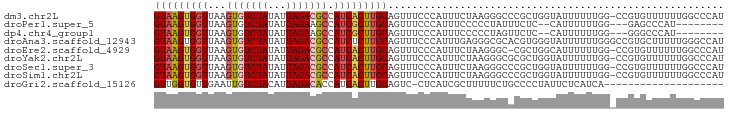
>dm3.chr2L 9369884 94 + 23011544 GUAAGUGGUUAAGUGUCUAUAUUAGACGCCAUCACUUGCAGUUUCCCAUUUCUAAGGGCCCGCUGGUAUUUUUUGG-CCGUGUUUUUUGGCCCAU (((((((((...(((((((...))))))).)))))))))................((((((((.(((........)-)))))......))))).. ( -31.80, z-score = -3.55, R) >droPer1.super_5 4274385 82 + 6813705 GUAAGUGGUUAAGUGUCUAUAUUAGAAGCCAUCGCUUGCAGUUUCCCAUUUCCCCCUAUUUCUC--CAUUUUUUGG---GAGCCCAU-------- (((((((((...((.((((...)))).)).)))))))))...((((((................--.......)))---))).....-------- ( -17.60, z-score = -2.08, R) >dp4.chr4_group1 4344542 82 - 5278887 GUAAGUGGUUAAGUGUCUAUAUUAGAAGCCAUCGCUUGCAGUUUCCCAUUUCCCCCUAGUUCUC--CAUUUUUUGG---GGGCCCAU-------- (((((((((...((.((((...)))).)).))))))))).............(((((((.....--......))))---))).....-------- ( -20.80, z-score = -1.94, R) >droAna3.scaffold_12943 722738 95 + 5039921 GUAAGUGGUUAAGUGUCUAUAUUAGACGCCAUCUCUUGCAGUUUCCCAUUUGAGGGCGCACGUGGGUAUUUUUUGGGCCGUGCUUUUUGGGCCAU (((((.(((...(((((((...))))))).))).))))).....(((((.((......)).))))).......(((.(((.......))).))). ( -28.50, z-score = -1.08, R) >droEre2.scaffold_4929 9982190 93 + 26641161 GUAAGUGGUUAAGUGUCUAUAUUAGACGCCAUCACUUGCAGUUUCCCAUUUCUAAGGGC-CGCUGGCAUUUUUUGG-CCGUGUUUUUUGGCCCAU (((((((((...(((((((...))))))).)))))))))................((((-(((((((........)-))).)).....))))).. ( -33.10, z-score = -3.82, R) >droYak2.chr2L 12042616 94 + 22324452 GUAAGUGGUUAAGUGUCUAUAUUAGACGCCAUCACUUGCAGUUUCCCAUUUCUAAGGGCGCGCUGGUAUUUUUUGG-CCGUGUUUUUUGGCCCAU (((((((((...(((((((...))))))).)))))))))................((((((((.(((........)-))))))......)))).. ( -30.50, z-score = -2.91, R) >droSec1.super_3 4827250 94 + 7220098 GUAAGUGGUUAAGUGUCUAUAUUAGACGCCAUCACUUGCAGUUUCCCAUUUCUAAGGGCCCGCUGGUAUUUUUUGG-CCGUGUUUUUUGGCCCAU (((((((((...(((((((...))))))).)))))))))................((((((((.(((........)-)))))......))))).. ( -31.80, z-score = -3.55, R) >droSim1.chr2L 9147330 94 + 22036055 GUAAGUGGUUAAGUGUCUAUAUUAGACGCCAUCACUUGCAGUUUCCCAUUUCUAAGGGCCCGCUGGUAUUUUUUGG-CCGUGUUUUUUGGCCCAU (((((((((...(((((((...))))))).)))))))))................((((((((.(((........)-)))))......))))).. ( -31.80, z-score = -3.55, R) >droGri2.scaffold_15126 6214244 74 - 8399593 GUUGGUGUUGAAUUGUCUACAUUAGACACCAUCACUUGCAGUC-CUCAUCGCUUUUUCUGCCCCUAUUCUCAUCA-------------------- (.(((((((....((....))...))))))).)....((((..-.............))))..............-------------------- ( -12.96, z-score = -1.73, R) >consensus GUAAGUGGUUAAGUGUCUAUAUUAGACGCCAUCACUUGCAGUUUCCCAUUUCUAAGGGCCCGCUGGUAUUUUUUGG_CCGUGUUUUUUGGCCCAU (((((((((...(((((((...))))))).)))))))))........................................................ (-15.18 = -15.50 + 0.32)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:28:04 2011