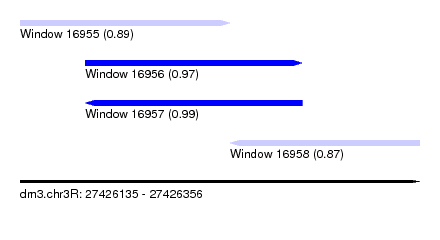
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 27,426,135 – 27,426,356 |
| Length | 221 |
| Max. P | 0.990093 |

| Location | 27,426,135 – 27,426,251 |
|---|---|
| Length | 116 |
| Sequences | 13 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.09 |
| Shannon entropy | 0.26308 |
| G+C content | 0.60528 |
| Mean single sequence MFE | -49.20 |
| Consensus MFE | -36.87 |
| Energy contribution | -36.58 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.890556 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 27426135 116 + 27905053 CGUUGGGGAUGCC----UUCCUCAAUGGUGGGCAGGUCGGAGGAGAAGCGGUUGAUCGGGCGCAAGACGCGCAGCGUGCGACUGGACAGCUUCCAGACAAGCCGCUGCUUGUAGGGCGCC (((((((((....----.))))))))).......((((((..(.....)..)))))).(((((...(((.((((((.((..(((((.....)))))....)))))))).)))...))))) ( -49.60, z-score = -0.92, R) >anoGam1.chr2R 9046403 120 + 62725911 CCAUGGGUAUGGUGGUGUUCGGCAAUCGUUGGCAAAUAGGAGGUCAACUUGUCAGUAGGGCGCAGCACGCGCAUAGUACGGGACGAAAGCUUCCAUACCAGCCGCUGCUUGUACGGGAUC (((..(((((((((((((..(((((..((((((.........))))))))))).(((..((((.....))))....)))(((((....).)))))))))).))).))))..)..)).... ( -38.90, z-score = -0.11, R) >droGri2.scaffold_14906 11700140 116 - 14172833 CAUUCGAUAUGCC----CUCCUCGAUGGUGGGCAGAUCGGAUGAGAAGCGAUUGAUCGGGCGCAGAACGCGCAGCGUGCGACUGGAUAACUUCCAGACAAGCCGCUGCUUGUAGGGCGCC ((((((((.((((----(.((.....)).))))).)))))))).....(((....)))(((((...(((.((((((.((..(((((.....)))))....)))))))).)))...))))) ( -57.60, z-score = -4.74, R) >droMoj3.scaffold_6540 26454903 116 - 34148556 CAUUCGAUAUGCC----CUCCUCAAUGGUGGGCAAAUCGGAUGAGAAACGAUUGAUCGGGCGCAGGACGCGCAGCGUGCGACUGGAUAAUUUCCAGACAAGCCGCUGCUUGUAGGGCGCC ((((((((.((((----(.((.....)).))))).)))))))).....(((....)))(((((...(((.((((((.((..(((((.....)))))....)))))))).)))...))))) ( -59.00, z-score = -5.82, R) >droVir3.scaffold_13047 9637639 116 - 19223366 CAUUCGAUAUACC----CUCCUCGAUGGUGGGCAAAUCGGAUGAGAAGCGAUUGAUCGGGCGCAGGACGCGCAGCGUGCGACUGGAUAAUUUCCAGACAAGCCGCUGCUUGUAGGGCGCC ((((((((...((----(.((.....)).)))...)))))))).....(((....)))(((((...(((.((((((.((..(((((.....)))))....)))))))).)))...))))) ( -52.20, z-score = -3.71, R) >droWil1.scaffold_181089 2956571 116 + 12369635 CAUUCGAGAUGCC----UUCCUCAAUGGUGGGCAGAUCCGAGGAGAAACGAUUGAUAGGGCGCAACACACGCAGCGUGCGACUGGAUAACUUCCAGACAAGCCGUUGCUUAUAGGGCGCC ((.(((...((((----(.((.....)).)))))..((....))....))).))....(((((.......((((((.((..(((((.....)))))....)))))))).......))))) ( -42.34, z-score = -2.45, R) >droSim1.chr3R_random 1288702 116 + 1307089 CGUUGGAGAUGCC----UUCCUCAAUGGUGGGCAGGUCGGAGGAGAAGCGGUUGAUCGGGCGCAAGACGCGCAGCGUGCGACUGGACAGCUUCCAGACAAGCCGCUGCUUGUAGGGCGCC (.((.((..((((----(.((.....)).)))))..)).)).).....(((....)))(((((...(((.((((((.((..(((((.....)))))....)))))))).)))...))))) ( -49.00, z-score = -0.94, R) >droSec1.super_31 86574 116 + 568655 CGUUGGAGAUGCC----UUCCUCAAUGGUGGGCAGGUCGGAGGAGAAGCGGUUGAUCGGGCGCAAGACGCGCAGCGUGCGACUGGACAGCUUCCAGACAAGCCGCUGCUUGUAGGGCGCC (.((.((..((((----(.((.....)).)))))..)).)).).....(((....)))(((((...(((.((((((.((..(((((.....)))))....)))))))).)))...))))) ( -49.00, z-score = -0.94, R) >droYak2.chr3R 28362078 116 + 28832112 CAUUGGAGAUGCC----UUCCUCAAUGGUGGGCAGGUCGGAGGAGAAGCGGUUGAUCGGGCGCAAGACGCGCAGCGUGCGACUGGACAGCUUCCAGACAAGCCGCUGCUUGUAGGGCGCC .........((((----(.((.....)).)))))((((((..(.....)..)))))).(((((...(((.((((((.((..(((((.....)))))....)))))))).)))...))))) ( -47.80, z-score = -0.94, R) >droEre2.scaffold_4820 9920613 116 - 10470090 CAUUGGGGAUGCC----UUCCUCAAUGGUGGGCAGGUCGGAGGAGAAGCGGUUGAUCGGGCGCAAGACGCGCAGCGUGCGACUGGACAGCUUCCAGACUAGCCGCUGCUUGUAGGGCGCC (((((((((....----.)))))))))...(((.(((((((((.....((((((..((.((((.....))))..))..)))))).....))))).)))).(((.(((....))))))))) ( -50.50, z-score = -1.36, R) >droAna3.scaffold_12911 1678902 116 - 5364042 CGUUGGAGAUGCC----UUCCUCAAUGGUGGGCAGGUCGGAUGAGAAGCGGUUGAUCGGGCGCAGGACGCGCAGCGUGCGACUGGAGAGCUUCCAGACUAGCCGCUGCUUGUAGGGCGCC ((((.((..((((----(.((.....)).)))))..)).)))).....(((....)))(((((...(((.((((((.((..((((((...))))))....)))))))).)))...))))) ( -50.60, z-score = -1.25, R) >dp4.chr2 22428012 116 - 30794189 CAUUGGAAAUGCC----UUCCUCAAUGGUGGGCAGGUCGGAGGAGAAGCGAUUGAUCGGGCGCAAGACGCGCAGCGUGCGACUGGACAACUUCCAGACAAGCCGUUGCUUGUAGGGCGCC ..((.((..((((----(.((.....)).)))))..)).)).......(((....)))(((((...(((.((((((.((..(((((.....)))))....)))))))).)))...))))) ( -46.50, z-score = -1.76, R) >droPer1.super_0 9622328 116 - 11822988 CAUUGGAAAUGCC----UUCCUCAAUGGUGGGCAGGUCGGAGGAGAAGCGAUUGAUCGGGCGCAAGACGCGCAGCGUGCGACUGGACAACUUCCAGACAAGCCGUUGCUUGUAGGGCGCC ..((.((..((((----(.((.....)).)))))..)).)).......(((....)))(((((...(((.((((((.((..(((((.....)))))....)))))))).)))...))))) ( -46.50, z-score = -1.76, R) >consensus CAUUGGAGAUGCC____UUCCUCAAUGGUGGGCAGGUCGGAGGAGAAGCGGUUGAUCGGGCGCAAGACGCGCAGCGUGCGACUGGACAGCUUCCAGACAAGCCGCUGCUUGUAGGGCGCC (.((.((..((((......((.....))..))))..)).)).).....(((....)))(((((...(((.((((((.((..(((((.....)))))....)))))))).)))...))))) (-36.87 = -36.58 + -0.28)

| Location | 27,426,171 – 27,426,291 |
|---|---|
| Length | 120 |
| Sequences | 13 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.46 |
| Shannon entropy | 0.26230 |
| G+C content | 0.56731 |
| Mean single sequence MFE | -48.44 |
| Consensus MFE | -37.42 |
| Energy contribution | -37.34 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.85 |
| SVM RNA-class probability | 0.971617 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 27426171 120 + 27905053 AGGAGAAGCGGUUGAUCGGGCGCAAGACGCGCAGCGUGCGACUGGACAGCUUCCAGACAAGCCGCUGCUUGUAGGGCGCCUUGUUCUGCCCGUUGUAAUGGGACAGAAUACCCGAAACGA ........(((......((((((...(((.((((((.((..(((((.....)))))....)))))))).)))...))))))((((((((((((....))))).))))))).)))...... ( -52.50, z-score = -2.80, R) >anoGam1.chr2R 9046443 120 + 62725911 AGGUCAACUUGUCAGUAGGGCGCAGCACGCGCAUAGUACGGGACGAAAGCUUCCAUACCAGCCGCUGCUUGUACGGGAUCUUGUGCUGACCGUUGUACAGCGAAAGAAUCUUCGUCACAA .((((..............((((.....))))..((((((((((....).((((.(((.(((....))).))).))))))))))))))))).((((...(((((......))))).)))) ( -36.90, z-score = 0.06, R) >droGri2.scaffold_14906 11700176 120 - 14172833 AUGAGAAGCGAUUGAUCGGGCGCAGAACGCGCAGCGUGCGACUGGAUAACUUCCAGACAAGCCGCUGCUUGUAGGGCGCCUUAUUCUGGCCAUUGUAAUGGGACAGAAUGCCCGACACAA ............((.((((((((...(((.((((((.((..(((((.....)))))....)))))))).)))...)).....((((((.((((....))))..))))))))))))))... ( -48.10, z-score = -3.09, R) >droMoj3.scaffold_6540 26454939 120 - 34148556 AUGAGAAACGAUUGAUCGGGCGCAGGACGCGCAGCGUGCGACUGGAUAAUUUCCAGACAAGCCGCUGCUUGUAGGGCGCCUUAUUCUGUCCAUUGUAAUGAGACAGAAUGCCCGAGACAA ...........(((.((((((((...(((.((((((.((..(((((.....)))))....)))))))).)))...)).....(((((((((((....))).))))))))))))))..))) ( -51.10, z-score = -4.77, R) >droVir3.scaffold_13047 9637675 120 - 19223366 AUGAGAAGCGAUUGAUCGGGCGCAGGACGCGCAGCGUGCGACUGGAUAAUUUCCAGACAAGCCGCUGCUUGUAGGGCGCCUUAUUCUGUCCAUUGUAGUGGGACAGAAUGCCCGAGACAA ...........(((.((((((((...(((.((((((.((..(((((.....)))))....)))))))).)))...)).....(((((((((.........)))))))))))))))..))) ( -52.00, z-score = -3.83, R) >droWil1.scaffold_181089 2956607 120 + 12369635 AGGAGAAACGAUUGAUAGGGCGCAACACACGCAGCGUGCGACUGGAUAACUUCCAGACAAGCCGUUGCUUAUAGGGCGCCUUAUUCUGGCCAUUAUAAUGGGAUAGAAUGCCCGAGACAA .((............((((((((.......((((((.((..(((((.....)))))....)))))))).......))))))))(((((.((((....))))..)))))...))....... ( -40.94, z-score = -3.09, R) >droSim1.chr3R_random 1288738 120 + 1307089 AGGAGAAGCGGUUGAUCGGGCGCAAGACGCGCAGCGUGCGACUGGACAGCUUCCAGACAAGCCGCUGCUUGUAGGGCGCCUUGUUCUGCCCGUUGUAGUGGGACAGAAUACCCGAAACGA ........(((......((((((...(((.((((((.((..(((((.....)))))....)))))))).)))...))))))(((((((((((......)))).))))))).)))...... ( -51.40, z-score = -2.10, R) >droSec1.super_31 86610 120 + 568655 AGGAGAAGCGGUUGAUCGGGCGCAAGACGCGCAGCGUGCGACUGGACAGCUUCCAGACAAGCCGCUGCUUGUAGGGCGCCUUGUUCUGCCCGUUGUAGUGGGACAGAAUACCCGAAACGA ........(((......((((((...(((.((((((.((..(((((.....)))))....)))))))).)))...))))))(((((((((((......)))).))))))).)))...... ( -51.40, z-score = -2.10, R) >droYak2.chr3R 28362114 120 + 28832112 AGGAGAAGCGGUUGAUCGGGCGCAAGACGCGCAGCGUGCGACUGGACAGCUUCCAGACAAGCCGCUGCUUGUAGGGCGCCUUGUUCUGCCCGUUGUAGUGGGACAGAAUACCCGAAACGA ........(((......((((((...(((.((((((.((..(((((.....)))))....)))))))).)))...))))))(((((((((((......)))).))))))).)))...... ( -51.40, z-score = -2.10, R) >droEre2.scaffold_4820 9920649 120 - 10470090 AGGAGAAGCGGUUGAUCGGGCGCAAGACGCGCAGCGUGCGACUGGACAGCUUCCAGACUAGCCGCUGCUUGUAGGGCGCCUUGUUCUGUCCGUUGUAGUGAGACAGAAUGCCCGAAACGA ..........(((..((((((((...(((.((((((.((..(((((.....)))))....)))))))).)))...)).....((((((((((......)).))))))))))))))))).. ( -49.00, z-score = -1.51, R) >droAna3.scaffold_12911 1678938 120 - 5364042 AUGAGAAGCGGUUGAUCGGGCGCAGGACGCGCAGCGUGCGACUGGAGAGCUUCCAGACUAGCCGCUGCUUGUAGGGCGCCUUGUUCUGCCCGUUGUAGUGGGACAGAAUGCCCGACACGA ........((((((...((((((...(((.((((((.((..((((((...))))))....)))))))).)))...)))))).((((((((((......)))).))))))...)))).)). ( -52.00, z-score = -1.64, R) >dp4.chr2 22428048 120 - 30794189 AGGAGAAGCGAUUGAUCGGGCGCAAGACGCGCAGCGUGCGACUGGACAACUUCCAGACAAGCCGUUGCUUGUAGGGCGCCUUGUUCUGACCAUUGUAGUGGGACAGAAUGCCCGAAACAA ...............((((((((...(((.((((((.((..(((((.....)))))....)))))))).)))...)).....((((((.((((....))))..))))))))))))..... ( -46.50, z-score = -2.36, R) >droPer1.super_0 9622364 120 - 11822988 AGGAGAAGCGAUUGAUCGGGCGCAAGACGCGCAGCGUGCGACUGGACAACUUCCAGACAAGCCGUUGCUUGUAGGGCGCCUUGUUCUGACCAUUGUAGUGGGACAGAAUGCCCGAAACAA ...............((((((((...(((.((((((.((..(((((.....)))))....)))))))).)))...)).....((((((.((((....))))..))))))))))))..... ( -46.50, z-score = -2.36, R) >consensus AGGAGAAGCGGUUGAUCGGGCGCAAGACGCGCAGCGUGCGACUGGACAGCUUCCAGACAAGCCGCUGCUUGUAGGGCGCCUUGUUCUGCCCGUUGUAGUGGGACAGAAUGCCCGAAACAA ...............((((((((...(((.((((((.((..(((((.....)))))....)))))))).)))...))))).(((((((.((((....))).).)))))))..)))..... (-37.42 = -37.34 + -0.08)


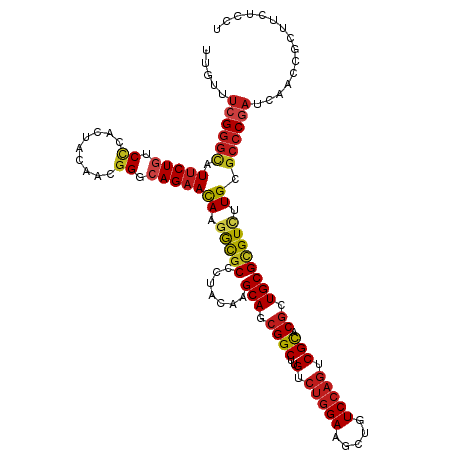
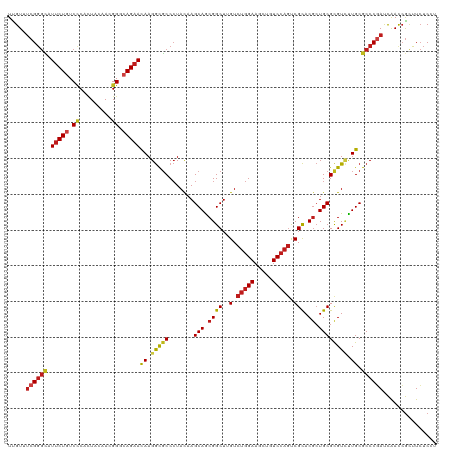
| Location | 27,426,171 – 27,426,291 |
|---|---|
| Length | 120 |
| Sequences | 13 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.46 |
| Shannon entropy | 0.26230 |
| G+C content | 0.56731 |
| Mean single sequence MFE | -46.75 |
| Consensus MFE | -30.09 |
| Energy contribution | -30.54 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.24 |
| Mean z-score | -3.25 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.40 |
| SVM RNA-class probability | 0.990093 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 27426171 120 - 27905053 UCGUUUCGGGUAUUCUGUCCCAUUACAACGGGCAGAACAAGGCGCCCUACAAGCAGCGGCUUGUCUGGAAGCUGUCCAGUCGCACGCUGCGCGUCUUGCGCCCGAUCAACCGCUUCUCCU ..(((((((((.(((((.(((........))))))))((((((((.......((((((((..(.(((((.....))))).))).)))))))))))))).))))))..))).......... ( -50.11, z-score = -3.50, R) >anoGam1.chr2R 9046443 120 - 62725911 UUGUGACGAAGAUUCUUUCGCUGUACAACGGUCAGCACAAGAUCCCGUACAAGCAGCGGCUGGUAUGGAAGCUUUCGUCCCGUACUAUGCGCGUGCUGCGCCCUACUGACAAGUUGACCU (((((.(((((....)))))...))))).(((((((..(((...((((((.(((....))).))))))...)))..(((..(((....((((.....))))..))).)))..))))))). ( -37.70, z-score = -0.69, R) >droGri2.scaffold_14906 11700176 120 + 14172833 UUGUGUCGGGCAUUCUGUCCCAUUACAAUGGCCAGAAUAAGGCGCCCUACAAGCAGCGGCUUGUCUGGAAGUUAUCCAGUCGCACGCUGCGCGUUCUGCGCCCGAUCAAUCGCUUCUCAU (((.(((((((((((((..((((....)))).)))))).....((...((..((((((((..(.(((((.....))))).))).))))))..))...))))))))))))........... ( -46.60, z-score = -3.15, R) >droMoj3.scaffold_6540 26454939 120 + 34148556 UUGUCUCGGGCAUUCUGUCUCAUUACAAUGGACAGAAUAAGGCGCCCUACAAGCAGCGGCUUGUCUGGAAAUUAUCCAGUCGCACGCUGCGCGUCCUGCGCCCGAUCAAUCGUUUCUCAU (((..((((((((((((((.(((....))))))))))).((((((.......((((((((..(.(((((.....))))).))).))))))))).)))..)))))).)))........... ( -49.41, z-score = -5.08, R) >droVir3.scaffold_13047 9637675 120 + 19223366 UUGUCUCGGGCAUUCUGUCCCACUACAAUGGACAGAAUAAGGCGCCCUACAAGCAGCGGCUUGUCUGGAAAUUAUCCAGUCGCACGCUGCGCGUCCUGCGCCCGAUCAAUCGCUUCUCAU (((..(((((((((((((((.........))))))))).((((((.......((((((((..(.(((((.....))))).))).))))))))).)))..)))))).)))........... ( -49.71, z-score = -4.82, R) >droWil1.scaffold_181089 2956607 120 - 12369635 UUGUCUCGGGCAUUCUAUCCCAUUAUAAUGGCCAGAAUAAGGCGCCCUAUAAGCAACGGCUUGUCUGGAAGUUAUCCAGUCGCACGCUGCGUGUGUUGCGCCCUAUCAAUCGUUUCUCCU ...(((.((.((((.((......)).)))).)))))(((.(((((..((((.(((.((((..(.(((((.....))))).))).)).))).))))..))))).))).............. ( -34.50, z-score = -1.37, R) >droSim1.chr3R_random 1288738 120 - 1307089 UCGUUUCGGGUAUUCUGUCCCACUACAACGGGCAGAACAAGGCGCCCUACAAGCAGCGGCUUGUCUGGAAGCUGUCCAGUCGCACGCUGCGCGUCUUGCGCCCGAUCAACCGCUUCUCCU ..(((((((((.(((((.(((........))))))))((((((((.......((((((((..(.(((((.....))))).))).)))))))))))))).))))))..))).......... ( -50.11, z-score = -3.44, R) >droSec1.super_31 86610 120 - 568655 UCGUUUCGGGUAUUCUGUCCCACUACAACGGGCAGAACAAGGCGCCCUACAAGCAGCGGCUUGUCUGGAAGCUGUCCAGUCGCACGCUGCGCGUCUUGCGCCCGAUCAACCGCUUCUCCU ..(((((((((.(((((.(((........))))))))((((((((.......((((((((..(.(((((.....))))).))).)))))))))))))).))))))..))).......... ( -50.11, z-score = -3.44, R) >droYak2.chr3R 28362114 120 - 28832112 UCGUUUCGGGUAUUCUGUCCCACUACAACGGGCAGAACAAGGCGCCCUACAAGCAGCGGCUUGUCUGGAAGCUGUCCAGUCGCACGCUGCGCGUCUUGCGCCCGAUCAACCGCUUCUCCU ..(((((((((.(((((.(((........))))))))((((((((.......((((((((..(.(((((.....))))).))).)))))))))))))).))))))..))).......... ( -50.11, z-score = -3.44, R) >droEre2.scaffold_4820 9920649 120 + 10470090 UCGUUUCGGGCAUUCUGUCUCACUACAACGGACAGAACAAGGCGCCCUACAAGCAGCGGCUAGUCUGGAAGCUGUCCAGUCGCACGCUGCGCGUCUUGCGCCCGAUCAACCGCUUCUCCU ..(((((((((.((((((((.........))))))))((((((((.......((((((((..(.(((((.....))))).))).)))))))))))))).))))))..))).......... ( -50.81, z-score = -4.04, R) >droAna3.scaffold_12911 1678938 120 + 5364042 UCGUGUCGGGCAUUCUGUCCCACUACAACGGGCAGAACAAGGCGCCCUACAAGCAGCGGCUAGUCUGGAAGCUCUCCAGUCGCACGCUGCGCGUCCUGCGCCCGAUCAACCGCUUCUCAU ....(((((((.(((((.(((........))))))))((.(((((.......((((((((..(.(((((.....))))).))).))))))))))).)).))))))).............. ( -49.41, z-score = -3.12, R) >dp4.chr2 22428048 120 + 30794189 UUGUUUCGGGCAUUCUGUCCCACUACAAUGGUCAGAACAAGGCGCCCUACAAGCAACGGCUUGUCUGGAAGUUGUCCAGUCGCACGCUGCGCGUCUUGCGCCCGAUCAAUCGCUUCUCCU (((..((((((.(((((.((.........)).)))))((((((((...((((((....))))))(((((.....)))))..((.....)))))))))).)))))).)))........... ( -44.60, z-score = -3.10, R) >droPer1.super_0 9622364 120 + 11822988 UUGUUUCGGGCAUUCUGUCCCACUACAAUGGUCAGAACAAGGCGCCCUACAAGCAACGGCUUGUCUGGAAGUUGUCCAGUCGCACGCUGCGCGUCUUGCGCCCGAUCAAUCGCUUCUCCU (((..((((((.(((((.((.........)).)))))((((((((...((((((....))))))(((((.....)))))..((.....)))))))))).)))))).)))........... ( -44.60, z-score = -3.10, R) >consensus UUGUUUCGGGCAUUCUGUCCCACUACAACGGGCAGAACAAGGCGCCCUACAAGCAGCGGCUUGUCUGGAAGCUGUCCAGUCGCACGCUGCGCGUCUUGCGCCCGAUCAACCGCUUCUCCU .....((((((.(((((.((.........)).)))))((.(((((.......(((.((((..(.(((((.....))))).))).)).)))))))).)).))))))............... (-30.09 = -30.54 + 0.45)

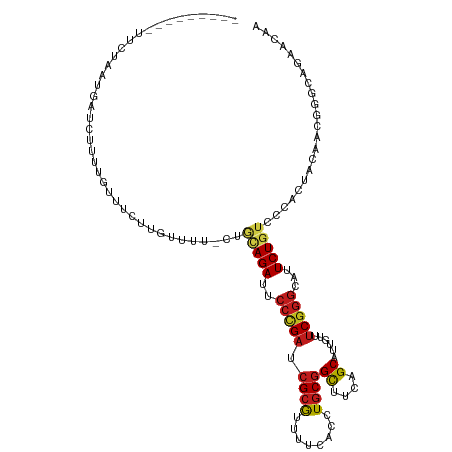
| Location | 27,426,251 – 27,426,356 |
|---|---|
| Length | 105 |
| Sequences | 13 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 71.53 |
| Shannon entropy | 0.62746 |
| G+C content | 0.46646 |
| Mean single sequence MFE | -24.32 |
| Consensus MFE | -13.72 |
| Energy contribution | -13.78 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.31 |
| Mean z-score | -0.89 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.871654 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 27426251 105 - 27905053 ---------CUCUAAUGUCCU--GGCUUCGUGUGCC-CCACAGAUUCCUGAUCGCGUUUUCACCUGCGGCUUCAGCAUCGUUUCGGGUAUUCUGUCCCAUUACAACGGGCAGAACAA ---------......(((.((--(.((...((((..-..(((((..(((((.((((........))))((....))......)))))...))))).....))))..)).))).))). ( -27.20, z-score = -0.53, R) >anoGam1.chr2R 9046523 108 - 62725911 ---------UUCUGCCCACUCUCUAUCGGCACGUUUGCUACAGAUCCCGGACAGCAGCUUCCUUUACAGUCACAGCAUUGUGACGAAGAUUCUUUCGCUGUACAACGGUCAGCACAA ---------...((((...........))))....((((...((((...((((((.............(((((......)))))((((....))))))))).)...))))))))... ( -25.50, z-score = -0.63, R) >droGri2.scaffold_14906 11700256 105 + 14172833 ---------AUUUAAUCAUCAUU--UUAUACAUUUU-CUGCAGAUUCCCGAUCGCGUUUUCACCUGCGGUUUUAGCAUUGUGUCGGGCAUUCUGUCCCAUUACAAUGGCCAGAAUAA ---------..............--.........((-((((........(((((((........)))))))....(((((((..((((.....))))...)))))))).)))))... ( -22.60, z-score = -1.44, R) >droMoj3.scaffold_6540 26455019 93 + 34148556 ---------------------AU--AUAUACAUCUA-UUGCAGAUACCCGAUCGCGUUUUUACCUGCGGCUUUAGCAUUGUCUCGGGCAUUCUGUCUCAUUACAAUGGACAGAAUAA ---------------------..--......((((.-....)))).(((((.((((........))))((....))......))))).((((((((.(((....))))))))))).. ( -24.30, z-score = -2.46, R) >droVir3.scaffold_13047 9637755 103 + 19223366 ---------AUAUAAUAAUUGUG--UUGUUCGCCU---UGCAGAUUCCCGAUCGCGUUUUCACCUGCGGUUUUAGCAUUGUCUCGGGCAUUCUGUCCCACUACAAUGGACAGAAUAA ---------..((((((....))--))))..((((---.((((......(((((((........)))))))......))))...))))(((((((((.........))))))))).. ( -25.50, z-score = -1.22, R) >droWil1.scaffold_181089 2956687 117 - 12369635 UCUUAAUUUGGGUUGCCUUUUCUCUUAUCUUCCAUGAUUCCAGAUUCCCGAUCGCGUUUUCACCUGCGGUUUCAGCAUUGUCUCGGGCAUUCUAUCCCAUUAUAAUGGCCAGAAUAA ......(((((..(((((..(((...(((......)))...))).....(((((((........))))))).............))))).......((((....))))))))).... ( -21.40, z-score = 0.37, R) >droSim1.chr3R_random 1288818 104 - 1307089 ----------UUCAAUGACCU--GGCUUCUUGUGCC-CCACAGAUUCCCGAUCGCGUUUUCACCUGCGGCUUCAGCAUCGUUUCGGGUAUUCUGUCCCACUACAACGGGCAGAACAA ----------.....((..((--(.((..(((((..-..(((((..(((((.((((........))))((....))......)))))...))))).....)))))..)))))..)). ( -27.40, z-score = -0.72, R) >droSec1.super_31 86690 105 - 568655 ---------UUCUAAUGGCCU--GUCUUCUUGUGCC-CCACAGAUUCCCGAUCGCGUUUUCACCUGCGGCUUCAGCAUCGUUUCGGGUAUUCUGUCCCACUACAACGGGCAGAACAA ---------......((..((--((((..(((((..-..(((((..(((((.((((........))))((....))......)))))...))))).....))))).))))))..)). ( -31.10, z-score = -2.10, R) >droYak2.chr3R 28362194 100 - 28832112 ---------UUCUAAU-------UGUUUCGUGUUCU-UCGCAGAUUCCUGAUCGCGUUUUCACCUGCGGCUUCAGCAUCGUUUCGGGUAUUCUGUCCCACUACAACGGGCAGAACAA ---------.......-------.......((((((-..(((((..(((((.((((........))))((....))......)))))...)))))(((........))).)))))). ( -25.30, z-score = -1.34, R) >droEre2.scaffold_4820 9920729 107 + 10470090 ---------UUCUAAUCUUCUUAUGUUUCGUGUUCC-CCGCAGAUUCCUGAUCGCGUUUUCACCUGCGGCUUCAGCAUCGUUUCGGGCAUUCUGUCUCACUACAACGGACAGAACAA ---------..........................(-(((..(((..(((((((((........)))))..)))).)))....))))..((((((((.........))))))))... ( -23.00, z-score = -0.71, R) >droAna3.scaffold_12911 1679018 101 + 5364042 ---------------CUAUAGCACCUCUUUUUCACGA-UCCAGAUUCCCGAUCGCGUUUUCACCUGCGGCUUCAGCAUCGUGUCGGGCAUUCUGUCCCACUACAACGGGCAGAACAA ---------------.....((.((.......(((((-(.............((((........))))((....))))))))..)))).(((((.(((........))))))))... ( -23.80, z-score = -0.78, R) >dp4.chr2 22428128 108 + 30794189 ---------UUCUAAUGAUUUUUUGUUGUCCCUUUUCUUGUAGAUUCCCGAUCGCGUUUUCACCUGCGGCUUUAGCAUUGUUUCGGGCAUUCUGUCCCACUACAAUGGUCAGAACAA ---------.((((..((.................))...))))..(((((.((((........))))((....))......)))))..(((((.((.........)).)))))... ( -19.63, z-score = 0.11, R) >droPer1.super_0 9622444 108 + 11822988 ---------UUAUAAUGAUUUUUUGUUAUCCCUUUUCUUGUAGAUUCCCGAUCGCGUUUUCACCUGCGGCUUUAGCAUUGUUUCGGGCAUUCUGUCCCACUACAAUGGUCAGAACAA ---------..((((..(....)..)))).................(((((.((((........))))((....))......)))))..(((((.((.........)).)))))... ( -19.40, z-score = -0.15, R) >consensus _________UUCUAAUGAUCUUUUGUUUCUUGUUUU_CUGCAGAUUCCCGAUCGCGUUUUCACCUGCGGCUUCAGCAUUGUUUCGGGCAUUCUGUCCCACUACAACGGGCAGAACAA .......................................(((((..(((((.((((........))))((....))......)))))...)))))...................... (-13.72 = -13.78 + 0.06)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:00:12 2011