
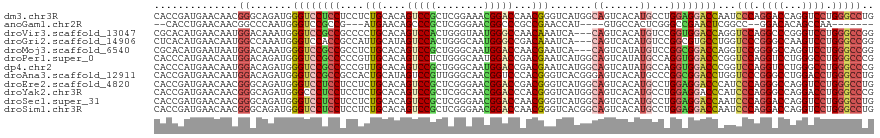
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 27,404,232 – 27,404,352 |
| Length | 120 |
| Max. P | 0.757073 |

| Location | 27,404,232 – 27,404,352 |
|---|---|
| Length | 120 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.77 |
| Shannon entropy | 0.49560 |
| G+C content | 0.64288 |
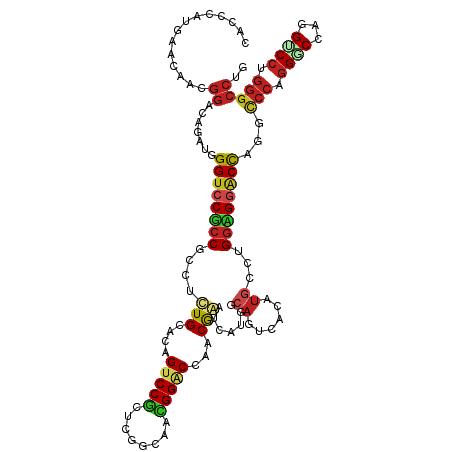
| Mean single sequence MFE | -53.15 |
| Consensus MFE | -23.52 |
| Energy contribution | -23.58 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.757073 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 27404232 120 - 27905053 CACCGAUGAACAACGGGCAGAUGGGUCCUCCUCCUCUGCACAGUCCGCUCGGAAACGGACCAACGGGUCAUGGCAGUCACAUGCCUGGAGGACCAAUCCCAGGACCAGGUCCUGGGCCUG .............(((((.(((.((((((((..................((....))((((....))))..((((......)))).)))))))).)))(((((((...)))))))))))) ( -52.40, z-score = -2.04, R) >anoGam1.chr2R 38441563 102 + 62725911 --CACCUGAACAACGGCCCAAUGGGUCCGCCG---AUGAACAGCCCGCUCGGGAACGGCCCGCCGAACCAU----GUGCCACUCGGGCCGAACUCGGCC--GGACACAGCCAA------- --............(((....((.((((((((---(.......(((....)))..(((((((.........----........)))))))...)))).)--)))).)))))..------- ( -37.33, z-score = -0.36, R) >droVir3.scaffold_13047 9616339 117 + 19223366 CGCACAUGAACAAUGGACAAAUGGGUCCGCCGCCCCUGCACAGUCCACUGGGUAAUGGGCCAACAAAUCA---CAGUCACAUGUCCGGUGGACCAGGUCCAGGCCCGGGUCCUGGGCCGG .............(((((.....((((((((((((.(((.(((....))).)))..)))).........(---((......)))..))))))))..)))))(((((((...))))))).. ( -49.50, z-score = -1.10, R) >droGri2.scaffold_14906 11679309 117 + 14172833 CUCACAUGAACAAUGGCCAAAUGGGUCCACCGCCAUUGCAUAGUCCACUGGGCAAUGGGCCGACAAAUCA---CAGUCACAUGUCCGGCGUGCCUGGUCCCGGGCCAAGUCCUGGGCCGG ...(((((......((((....((.....))..((((((.(((....))).))))))))))(((......---..))).)))))(((((..(((((....)))))((.....)).))))) ( -43.50, z-score = -0.22, R) >droMoj3.scaffold_6540 26434640 117 + 34148556 CGCACAUGAAUAAUGGACAAAUGGGUCCGCCGCCUCUGCACAGUCCGCUGGGCAAUGGACCAACGAAUCA---CAGUCAUAUGUCCGGCGGACCAGGUCCGGGGCCAGGUCCUGGGCCGG .(..(((.....)))..).....((((((((((((..((.......)).))))...((((....((....---...))....)))))))))))).((((((((((...)))))))))).. ( -50.60, z-score = -1.72, R) >droPer1.super_0 9600352 120 + 11822988 CACCCAUGAACAAUGGACAGAUGGGUCCGCCCCCGUUGCACAGUCCUCUGGGCAAUGGACCGACGAAUCAUGGCAGUCAUAUGCCAGGUGGACCCGGUCCAGGUCCUGGGCCUGGGCCCG ...((((.....))))......(((((((((.(((((((.(((....))).)))))))............(((((......))))))))))))))(((((((((.....))))))))).. ( -62.10, z-score = -3.56, R) >dp4.chr2 22406177 120 + 30794189 CACCCAUGAACAAUGGACAGAUGGGUCCGCCCCCGUUGCACAGUCCGCUGGGCAAUGGACCGACGAAUCAUGGCAGUCAUAUGCCAGGUGGACCCGGUCCAGGUCCUGGGCCUGGGCCCG ...((((.....))))......(((((((((.(((((((.(((....))).)))))))............(((((......))))))))))))))(((((((((.....))))))))).. ( -62.80, z-score = -3.59, R) >droAna3.scaffold_12911 1657380 120 + 5364042 CACCGAUGAACAAUGGACAGAUGGGUCCGCCGCCACUGCAUAGUCCGUUGGGCAACGGUCCCACGGGUCACGGGAGUCACAUGCCCGGCGGACCUGGUCCCGGGCCUGGACCUGGGCCUG ..............((((....((((((((((.....((((...((((.((((....)))).))))((.((....)).)))))).)))))))))).))))(((((((......))))))) ( -54.10, z-score = -0.34, R) >droEre2.scaffold_4820 9898532 120 + 10470090 CACCGAUGAACAACGGGCAGAUGGGUCCUCCUCCUCUGCACAGUCCGCUCGGGAACGGACCGACGGGUCAUGGCAGUCACAUGCCUGGAGGACCCAUCCCAGGGCCAGGUCCUGGGCCUG .............(((((.((((((((((((((((..((.......))..))))...((((....))))..((((......)))).))))))))))))(((((((...)))))))))))) ( -60.90, z-score = -2.87, R) >droYak2.chr3R 28339229 120 - 28832112 CACCGAUGAACAACGGGCAGAUGGGCCCUCCUCCUCUGCACAGUCCGCUCGGCAACGGACCCACGGGUCAUGGCAGUCACAUGCCUGGAGGACCCAUCCCAGGGCCAGGACCUGGGCCCG .............(((((.((((((.(((((..................((....))((((....))))..((((......)))).))))).))))))(((((.......)))))))))) ( -54.70, z-score = -1.72, R) >droSec1.super_31 64699 120 - 568655 CACCGAUGAACAACGGGCAGAUGGGUCCUCCUCCUCUGCACAGUCCGCUCGGGAACGGACCAACGGGUCAUGGCAGUCACAUGCCUGGAGGACCAAUCCCAGGACCAGGUCCUGGGCCUG .............(((((.(((.((((((((((((..((.......))..))))...((((....))))..((((......)))).)))))))).)))(((((((...)))))))))))) ( -54.80, z-score = -2.25, R) >droSim1.chr3R 27040102 120 - 27517382 CACCGAUGAACAACGGGCAGAUGGGUCCUCCUCCUCUGCACAGUCCGCUCGGGAACGGACCAACGGGUCACGGCAGUCACAUGCCUGGAGGACCAAUCCCAGGACCAGGUCCUGGGCCUG .............(((((.(((.((((((((((((..((.......))..))))...((((....))))..((((......)))).)))))))).)))(((((((...)))))))))))) ( -55.10, z-score = -2.38, R) >consensus CACCCAUGAACAACGGACAGAUGGGUCCGCCGCCUCUGCACAGUCCGCUCGGCAACGGACCAACGAAUCAUGGCAGUCACAUGCCUGGAGGACCAGGCCCAGGGCCAGGUCCUGGGCCUG ..............((.......((((((((....(((....(((((........)))))...))).......((......))...))))))))...(((.((((...)))).))))).. (-23.52 = -23.58 + 0.06)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:00:06 2011