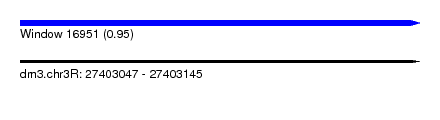
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 27,403,047 – 27,403,145 |
| Length | 98 |
| Max. P | 0.946905 |

| Location | 27,403,047 – 27,403,145 |
|---|---|
| Length | 98 |
| Sequences | 15 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.22 |
| Shannon entropy | 0.35347 |
| G+C content | 0.51584 |
| Mean single sequence MFE | -35.98 |
| Consensus MFE | -17.49 |
| Energy contribution | -16.30 |
| Covariance contribution | -1.19 |
| Combinations/Pair | 1.54 |
| Mean z-score | -2.82 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.52 |
| SVM RNA-class probability | 0.946905 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
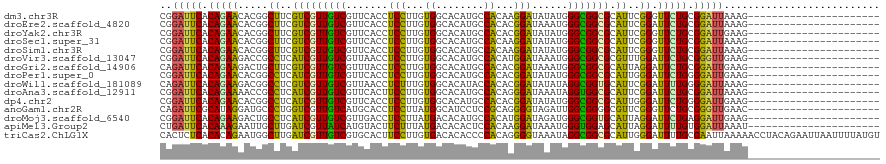
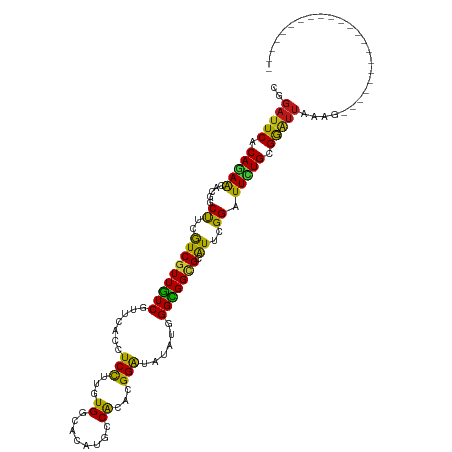
>dm3.chr3R 27403047 98 + 27905053 CGGAUUCACAGAACACGGCUUCGUCGUUGUCGUUCACCUCCUUGUGGCACAUGCCACAAGGAUAUAUGGGCGGCGCAUUCGGGUUCUGCGGAUUAAAG---------------------- ..(((((.(((((((((((...))))).((((((((..((((((((((....))))))))))....))))))))........)))))).)))))....---------------------- ( -44.10, z-score = -5.34, R) >droEre2.scaffold_4820 9897353 98 - 10470090 CGGAUUCACAGAACACGGCUUCGUCGUUGUCGUUCACCUCCUUGUGGCACAUGCCACACGGAUAAAUGGGCGGCGCAUUCGGAUUCUGCGGAUUAAAG---------------------- ..(((((.(((((..(((...((((((..(((((....(((.((((((....)))))).)))..)))))))))))...)))..))))).)))))....---------------------- ( -37.20, z-score = -3.17, R) >droYak2.chr3R 28338047 98 + 28832112 CGGAUUCACAGAACACGGCUUCGUCGUUGUCGUUCACCUCCUUGUGGCACAUGCCACACGGAUAUAUGGGCGGCGCAUUCGGGUUCUGCGGAUUGAAG---------------------- ..(((((.(((((((((((...))))).((((((((..(((.((((((....)))))).)))....))))))))........)))))).)))))....---------------------- ( -40.00, z-score = -3.31, R) >droSec1.super_31 63513 98 + 568655 CGGAUUCACAGAACACGGCUUCGUCGUUGUCGUUCACCUCCUUGUGGCACAUGCCACAAGGAUAUAUGGGCGGCGCAUUCGGGUUCUGCGGAUUAAAG---------------------- ..(((((.(((((((((((...))))).((((((((..((((((((((....))))))))))....))))))))........)))))).)))))....---------------------- ( -44.10, z-score = -5.34, R) >droSim1.chr3R 27038917 98 + 27517382 CGGAUUCACAGAACACGGCUUCGUCGUUGUCGUUCACCUCCUUGUGGCACAUGCCACAAGGAUAUAUGGGCGGCGCAUUCGGGUUCUGCGGAUUAAAG---------------------- ..(((((.(((((((((((...))))).((((((((..((((((((((....))))))))))....))))))))........)))))).)))))....---------------------- ( -44.10, z-score = -5.34, R) >droVir3.scaffold_13047 9614914 98 - 19223366 CGGAUUCACAGAAGACCGCCUCAUCGUUGUCGUUAACCUCCUUGUGGCACAUGCCACAUGGAUAAAUGGGCGGCGCGUUUGGAUUCUGCGGGUUGAAG---------------------- ..(((((.(((((.....((....(((.((((((....(((.((((((....)))))).)))......)))))))))...)).))))).)))))....---------------------- ( -34.70, z-score = -1.55, R) >droGri2.scaffold_14906 11677896 98 - 14172833 CAGAUUCACAGAAGACUGCUUCGUCGUUGUCGUUUACCUCCUUGUGGCACAUGCCACACGGAUAAAUGGGCGGCGCAUUAGGAUUCUGCGGAUUGAAG---------------------- ..(((((.(((((..(((...((((((..(((((((..(((.((((((....)))))).))))))))))))))))...)))..))))).)))))....---------------------- ( -40.10, z-score = -4.32, R) >droPer1.super_0 9599038 98 - 11822988 CGGAUUCACAGAACACGGCCUCAUCGUUGUCGUUCACCUCCUUGUGGCACAUGCCACACGGAUAUAUGGGCGGCGCAUUGGGAUUCUGGGGAUUGAAG---------------------- ..(((((.(((((.....((.((..((.((((((((..(((.((((((....)))))).)))....))))))))))..)))).))))).)))))....---------------------- ( -35.50, z-score = -2.08, R) >droWil1.scaffold_181089 2934976 98 + 12369635 CAGAUUCACAGAAGACGGCCUCGUCGUUGUCGUUAACCUCUUUGUGGCACAUACCACACGGAUAUAUAGGCGGUGCAUUCGGAUUUUGGGGAUUAAAG---------------------- ..(((((.(((((..((((.(((((((((....)))).(((.(((((......))))).)))......))))).))...))..))))).)))))....---------------------- ( -27.10, z-score = -1.36, R) >droAna3.scaffold_12911 1656195 98 - 5364042 CGGAUUCACAGAAAACCGCCUCAUCGUUGUCGUUCACUUCCUUGUGGCACAUGCCACAGGGAUAAAUAGGUGGCGCAUUCGGAUUCUGCGGAUUAAAG---------------------- ..(((((.(((((..(((......(((..((.......((((((((((....))))))))))......))..)))....))).))))).)))))....---------------------- ( -37.12, z-score = -3.95, R) >dp4.chr2 22404860 98 - 30794189 CGGAUUCACAGAACACGGCCUCAUCGUUGUCGUUCACCUCCUUGUGGCACAUGCCACACGGAUAUAUGGGCGGCGCAUUGGGAUUCUGGGGAUUGAAG---------------------- ..(((((.(((((.....((.((..((.((((((((..(((.((((((....)))))).)))....))))))))))..)))).))))).)))))....---------------------- ( -35.50, z-score = -2.08, R) >anoGam1.chr2R 38440263 98 - 62725911 CAGAUUCGCAUAGGAUGCCCUGGUCGUUGUCAUGCACCUCCUUAUGGCAUCCUCCGCAGGGGUAGAUUGGCGGGGCGUUCGGGUUCUGCGGGUUGAAC---------------------- ..((((((((..((((((((((.(.(((((((((........))))))(((((.....))))).))).).))))))))))......))))))))....---------------------- ( -34.10, z-score = 0.12, R) >droMoj3.scaffold_6540 26433302 98 - 34148556 CGGAUUCACAGAAGACUGCCUCAUCGUUGUCGUUGACCUCCUUAUGACACAUGCCACAUGGAUAGAUGGGCGGUGCAUUAGGAUUCUGAGGAUUGAAG---------------------- ..(((((.((((((((.((......)).))).......((((.(((.(((.((((.(((......))))))))))))).))))))))).)))))....---------------------- ( -30.00, z-score = -1.28, R) >apiMel3.Group2 13837433 98 - 14465785 CUGAUUCACAAAGAAUUGCUUGAUCGUUAUCAUGUACUUCUUUAUGACACACUCCACAAGGAUAAAUGGGUGGAGCAUUAGGAUUUUGUGGAUUAAAU---------------------- .((((((((((((((.(((.((((....)))).))).))))))........((((((..(......)..))))))............))))))))...---------------------- ( -24.90, z-score = -2.20, R) >triCas2.ChLG1X 168374 120 + 8109244 CACUCUCACACAGAAUGGCUUGAUCGUUGUCGUGCACUUCCUUGUGACACACCCCACAGGGGUAAAUAGGCGGCGCAUUGGGAUUUUGCGAAUUAAAAACCUACAGAAUUAAUUUUAUGU ........(.((((((..((..(((((((((((((((......))).)))(((((...))))).....))))))).))..)))))))).).............................. ( -31.20, z-score = -1.08, R) >consensus CGGAUUCACAGAACACGGCUUCGUCGUUGUCGUUCACCUCCUUGUGGCACAUGCCACACGGAUAUAUGGGCGGCGCAUUCGGAUUCUGCGGAUUAAAG______________________ ..(((((.(((((.....((..(((((((((.......(((...((........))...)))......))))))).))..)).))))).))))).......................... (-17.49 = -16.30 + -1.19)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:00:05 2011