| Sequence ID | dm3.chr3R |
|---|---|
| Location | 27,401,124 – 27,401,192 |
| Length | 68 |
| Max. P | 0.765162 |

| Location | 27,401,124 – 27,401,192 |
|---|---|
| Length | 68 |
| Sequences | 11 |
| Columns | 69 |
| Reading direction | forward |
| Mean pairwise identity | 63.98 |
| Shannon entropy | 0.78318 |
| G+C content | 0.39852 |
| Mean single sequence MFE | -10.35 |
| Consensus MFE | -5.03 |
| Energy contribution | -4.79 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 2.00 |
| Mean z-score | -0.42 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.765162 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
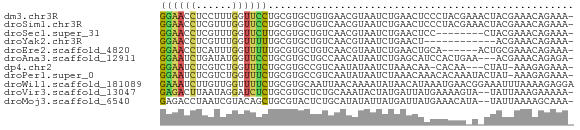
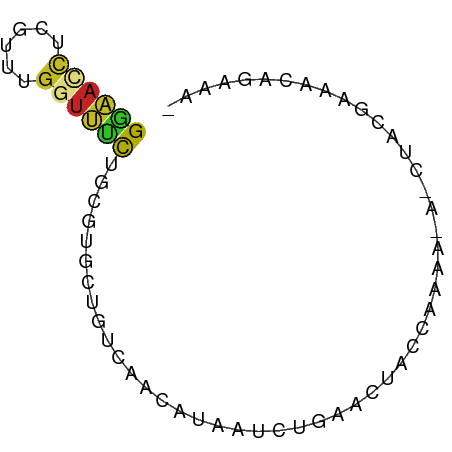
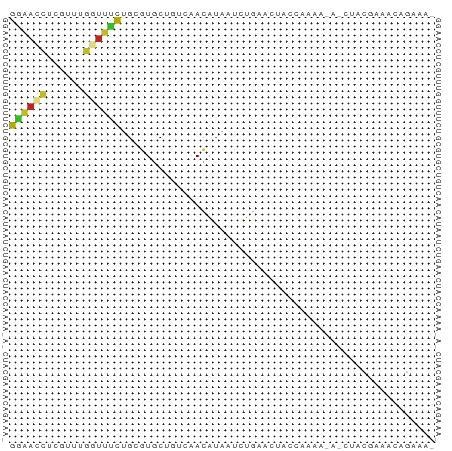
>dm3.chr3R 27401124 68 + 27905053 GGAACCUCCUUUGGUUCCUGCGUGCUGUGAACGUAAUCUGAACUCCCUACGAAACUACGAAACAGAAA- ((((((......))))))......((((...((((....................))))..))))...- ( -14.85, z-score = -2.30, R) >droSim1.chr3R 27036988 68 + 27517382 GGAACCUCGUUUGGUUCCUGCGUGCUGUCAACGUAAUCUGAACUCCCUACGAAACUACGAAACAGAAA- ((((((......))))))(((((.......))))).((((......................))))..- ( -14.85, z-score = -1.50, R) >droSec1.super_31 61606 60 + 568655 GGAACCUCGUUUGGUUCUUGCGUGCUGUCAACGUAAUCUGAACUCC--------CUACGAAACAGAAA- .(((((......)))))((((((.......))))))((((...((.--------....))..))))..- ( -13.20, z-score = -1.26, R) >droYak2.chr3R 28336188 56 + 28832112 GGAACCUCGUUUGGUUUUUGCGUGCUGUCAACGUAAUCUGAACU------------ACGAAACAGAAA- .(((((......)))))((((((.......))))))((((....------------......))))..- ( -10.90, z-score = -0.32, R) >droEre2.scaffold_4820 9895235 62 - 10470090 GGAACCUCAUUUGGUUUUUGCGUGCUGUCAACGUAAUCUGAACUGCA------ACUGCGAAACAGAAA- .(((((......)))))((((((.......))))))((((...(((.------...)))...))))..- ( -14.60, z-score = -1.18, R) >droAna3.scaffold_12911 1654456 65 - 5364042 GGAAUCUGAUAUGGUUCCUGCGUGCUGCCAACAUAAUCUGAGCAUCCACUGAA---ACGAAACAGAGA- ((((((......))))))((.(((((..............))))).))(((..---......)))...- ( -12.54, z-score = -0.29, R) >dp4.chr2 22402525 63 - 30794189 GGAAUCUCGUCUGGUUUCUGCGUGCCGUCAAUAUAAUCUAAACAA-CACAA---CUAU-AAAGAGAAA- ....((((...(((((.....(((..((.............))..-)))))---))).-...))))..- ( -5.32, z-score = 0.95, R) >droPer1.super_0 9596585 67 - 11822988 GGAAUCUCGUCUGGUUUCUGCGUGCCGUCAAUAUAAUCUAAACAAACACAAAUACUAU-AAAGAGAAA- ....((((...((((...((.((......................)).))...)))).-...))))..- ( -6.55, z-score = 0.30, R) >droWil1.scaffold_181089 2932794 69 + 12369635 GAAAUCUUGUUGGUUUUCUGCGUGCAAUUAACAAAAUAUAACAUAAAUGAACGGAAAUUUAAAAGAGGA ....((((...(((((((..(((.......................)))...)))))))...))))... ( -6.50, z-score = 0.57, R) >droVir3.scaffold_13047 9612234 66 - 19223366 GAGACUUAAUAGGAUCUCUGCGUGCUCUGCAAAUACUAUGAUUAUGAAAAGUA--UAUUAAAGAAAAA- ((((((.....)).))))((((.....)))).(((((.(........).))))--)............- ( -6.90, z-score = 0.34, R) >droMoj3.scaffold_6540 26430992 66 - 34148556 GAGACCUAAUCGUACAGCUGCGUACUCUGCAUAUAUUAUGAUUAUGAAACAUA--UAUUAAAAGCAAA- ....(.((((((((....((((.....)))).....)))))))).).......--.............- ( -7.60, z-score = 0.09, R) >consensus GGAACCUCGUUUGGUUUCUGCGUGCUGUCAACAUAAUCUGAACUACCAAAA_A_CUACGAAACAGAAA_ ((((((......))))))................................................... ( -5.03 = -4.79 + -0.24)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:00:05 2011