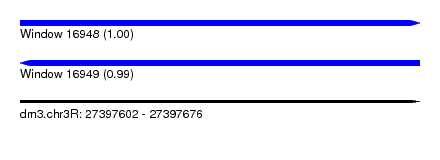
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 27,397,602 – 27,397,676 |
| Length | 74 |
| Max. P | 0.999564 |

| Location | 27,397,602 – 27,397,676 |
|---|---|
| Length | 74 |
| Sequences | 7 |
| Columns | 78 |
| Reading direction | forward |
| Mean pairwise identity | 62.19 |
| Shannon entropy | 0.77031 |
| G+C content | 0.35630 |
| Mean single sequence MFE | -14.57 |
| Consensus MFE | -8.61 |
| Energy contribution | -9.43 |
| Covariance contribution | 0.82 |
| Combinations/Pair | 1.53 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.02 |
| SVM RNA-class probability | 0.999564 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
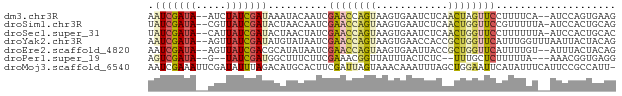
>dm3.chr3R 27397602 74 + 27905053 AAUCGAUA--AUCUAUCGAUAAAUACAAUCGAACCAGUAAGUGAAUCUCAACUAGUUCCUUUUCA--AUCCAGUGAAG .(((((((--...)))))))..........((((.(((.((.....))..))).))))...((((--......)))). ( -10.00, z-score = -1.24, R) >droSim1.chr3R 27033471 75 + 27517382 UAUCGAUA--CGUUAUCGAUACUAACAAUCGAACCAGUAAGUGAAUCUCAACUGGUUCCGUUUUUA-AUCCACUGCAG ((((((((--...)))))))).(((.(((.((((((((.((.....))..)))))))).))).)))-........... ( -17.50, z-score = -2.98, R) >droSec1.super_31 58081 75 + 568655 UAUCGAUA--CAUUAUCGAUACUAACUAUCGAACCAGUAAGUGAAUCUCAACUGGUUCCUUUUUUA-AUCCACUGCAC ((((((((--...)))))))).........((((((((.((.....))..))))))))........-........... ( -16.50, z-score = -3.66, R) >droYak2.chr3R 28332679 76 + 28832112 AAUCGAUA--AGUUAUCGAUAUGUAUAAUCGAACCAGUAAGUGAACCACCGCUGGUUCAUUUGGUUUAAUUACUACAG .(((((((--...))))))).((((((((..((((((...((((((((....))))))))))))))..)))).)))). ( -26.20, z-score = -5.65, R) >droEre2.scaffold_4820 9891717 74 - 10470090 AAUCGAUA--AGUUAUCGACGCAUAUAAUCGAACCAGUAAGUGAAUUACCGCUGGUUCAUUUUGU--AUUUACUACAG ..((((((--...))))))...((((((..((((((((..(((...))).))))))))...))))--))......... ( -16.40, z-score = -2.37, R) >droPer1.super_19 348833 69 + 1869541 AGUCGAUA--G--UAUCGAUGGCUUUCUUCGAAACGGUUAUUUACUCUC--UUUGCUCUUUUUA---AAACGGUGAGG ....((.(--(--((..((((((((((...)))..))))))))))).))--....(((......---.......))). ( -7.52, z-score = 1.80, R) >droMoj3.scaffold_6540 26427359 77 - 34148556 AAUCGAAAUUCGAUAUUUAGACAUGCACUUCGAUUAGUAAACAAAUUUAGCUGGAAUUCAUAUUUCAUUCCGCCAUU- (((((((.....................)))))))..............((.(((((.........)))))))....- ( -7.90, z-score = 0.30, R) >consensus AAUCGAUA__AGUUAUCGAUACAUACAAUCGAACCAGUAAGUGAAUCUCAACUGGUUCCUUUUUU__AUCCACUGCAG .(((((((.....)))))))..........((((((((............)))))))).................... ( -8.61 = -9.43 + 0.82)
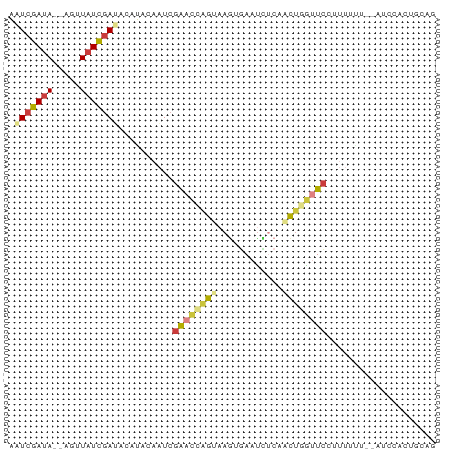
| Location | 27,397,602 – 27,397,676 |
|---|---|
| Length | 74 |
| Sequences | 7 |
| Columns | 78 |
| Reading direction | reverse |
| Mean pairwise identity | 62.19 |
| Shannon entropy | 0.77031 |
| G+C content | 0.35630 |
| Mean single sequence MFE | -16.61 |
| Consensus MFE | -6.79 |
| Energy contribution | -8.47 |
| Covariance contribution | 1.68 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.63 |
| SVM RNA-class probability | 0.993574 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
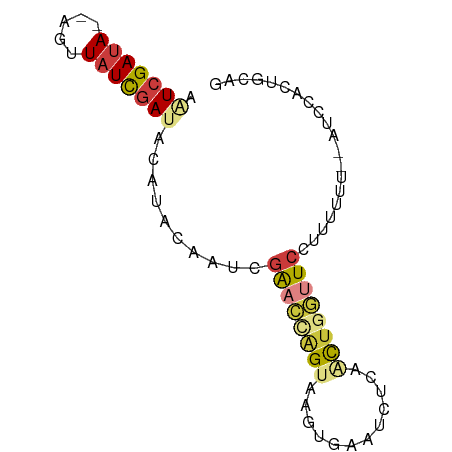
>dm3.chr3R 27397602 74 - 27905053 CUUCACUGGAU--UGAAAAGGAACUAGUUGAGAUUCACUUACUGGUUCGAUUGUAUUUAUCGAUAGAU--UAUCGAUU .((((......--))))...((((((((.(((.....)))))))))))..........(((((((...--))))))). ( -15.60, z-score = -1.01, R) >droSim1.chr3R 27033471 75 - 27517382 CUGCAGUGGAU-UAAAAACGGAACCAGUUGAGAUUCACUUACUGGUUCGAUUGUUAGUAUCGAUAACG--UAUCGAUA .........((-(((.....((((((((.(((.....))))))))))).....)))))(((((((...--))))))). ( -18.90, z-score = -1.70, R) >droSec1.super_31 58081 75 - 568655 GUGCAGUGGAU-UAAAAAAGGAACCAGUUGAGAUUCACUUACUGGUUCGAUAGUUAGUAUCGAUAAUG--UAUCGAUA .........((-(((.....((((((((.(((.....))))))))))).....)))))(((((((...--))))))). ( -18.90, z-score = -2.29, R) >droYak2.chr3R 28332679 76 - 28832112 CUGUAGUAAUUAAACCAAAUGAACCAGCGGUGGUUCACUUACUGGUUCGAUUAUACAUAUCGAUAACU--UAUCGAUU .((((.((((..(((((..(((((((....))))))).....)))))..)))))))).(((((((...--))))))). ( -22.20, z-score = -4.02, R) >droEre2.scaffold_4820 9891717 74 + 10470090 CUGUAGUAAAU--ACAAAAUGAACCAGCGGUAAUUCACUUACUGGUUCGAUUAUAUGCGUCGAUAACU--UAUCGAUU .((((.....)--)))...((((((((.(((.....)))..)))))))).........(((((((...--))))))). ( -16.90, z-score = -2.27, R) >droPer1.super_19 348833 69 - 1869541 CCUCACCGUUU---UAAAAAGAGCAAA--GAGAGUAAAUAACCGUUUCGAAGAAAGCCAUCGAUA--C--UAUCGACU .(((...((((---(....)))))...--)))(((..(((...((.((((.(.....).)))).)--)--)))..))) ( -8.10, z-score = -0.30, R) >droMoj3.scaffold_6540 26427359 77 + 34148556 -AAUGGCGGAAUGAAAUAUGAAUUCCAGCUAAAUUUGUUUACUAAUCGAAGUGCAUGUCUAAAUAUCGAAUUUCGAUU -..(((((((((.........))))).))))............(((((((((..((((....))))...))))))))) ( -15.70, z-score = -1.58, R) >consensus CUGCAGUGGAU__AAAAAAGGAACCAGUUGAGAUUCACUUACUGGUUCGAUUGUAUGUAUCGAUAACG__UAUCGAUU ....................(((((((..............)))))))..........(((((((.....))))))). ( -6.79 = -8.47 + 1.68)

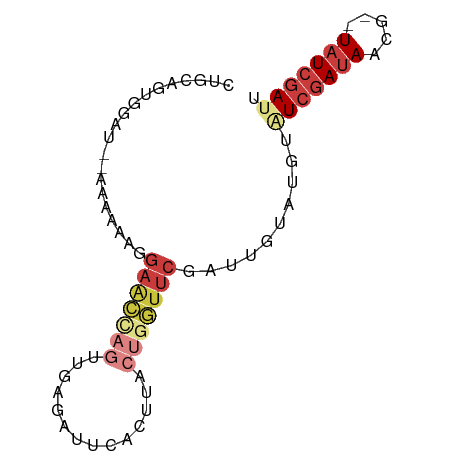
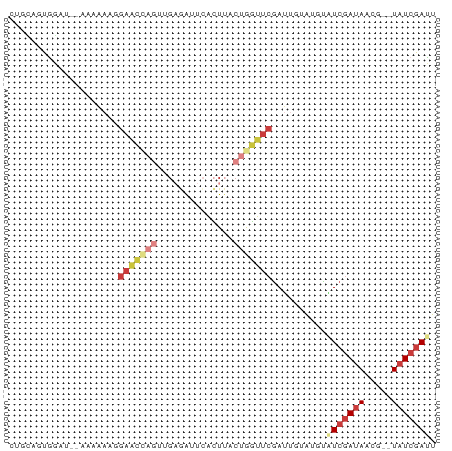
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:00:04 2011