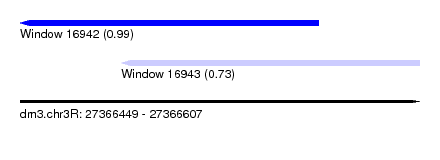
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 27,366,449 – 27,366,607 |
| Length | 158 |
| Max. P | 0.993512 |

| Location | 27,366,449 – 27,366,567 |
|---|---|
| Length | 118 |
| Sequences | 12 |
| Columns | 123 |
| Reading direction | reverse |
| Mean pairwise identity | 75.27 |
| Shannon entropy | 0.47449 |
| G+C content | 0.41258 |
| Mean single sequence MFE | -29.22 |
| Consensus MFE | -17.91 |
| Energy contribution | -18.06 |
| Covariance contribution | 0.15 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.62 |
| SVM RNA-class probability | 0.993512 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
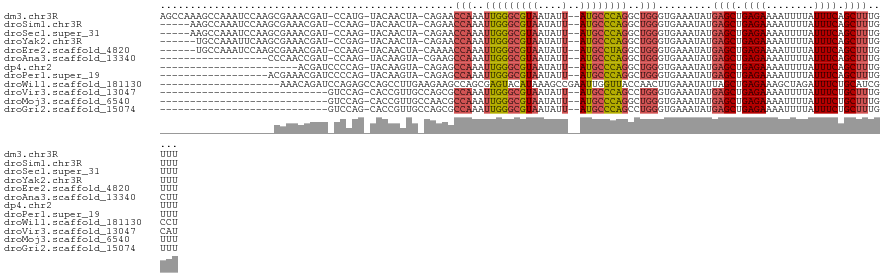
>dm3.chr3R 27366449 118 - 27905053 AGCCAAAGCCAAAUCCAAGCGAAACGAU-CCAUG-UACAACUA-CAGAACCAAAUUGGGCGUAAUAUU--AUGCCCAGGCUGGGUGAAAUAUGAGCUGAGAAAAUUUUAUUUCAGCUUUGUUU .(((..((((..................-...((-((....))-)).........((((((((....)--))))))))))).))).(((((.(((((((((........)))))))))))))) ( -33.00, z-score = -2.76, R) >droSim1.chr3R 27002416 113 - 27517382 -----AAGCCAAAUCCAAGCGAAACGAU-CCAAG-UACAACUA-CAGAACCAAAUUGGGCGUAAUAUU--AUGCCCAGGCUGGGUGAAAUAUGAGCUGAGAAAAUUUUAUUUCAGCUUUGUUU -----.......(((((.((........-....(-((....))-).........(((((((((....)--))))))))))))))).(((((.(((((((((........)))))))))))))) ( -30.60, z-score = -2.66, R) >droSec1.super_31 27140 113 - 568655 -----AAGCCAAAUCCAAGCGAAACGAU-CCAAG-UACAACUA-CAGAACCAAAUUGGGCGUAAUAUU--AUGCCCAGGCUGGGUGAAAUAUGAGCUGAGAAAAUUUUAUUUCAGCUUUGUUU -----.......(((((.((........-....(-((....))-).........(((((((((....)--))))))))))))))).(((((.(((((((((........)))))))))))))) ( -30.60, z-score = -2.66, R) >droYak2.chr3R 28301161 112 - 28832112 ------UGCCAAAUUCAAGCGAAACGAU-CCGAG-UACAACUA-CAGAACCAAAUUGGGCGUAAUAUU--AUGCCCAGGCUGGGUGAAAUAUGAGCUGAGAAAAUUUUAUUUCAGCUUUGUUU ------.......((((...........-....(-((....))-)....(((..(((((((((....)--))))))))..))).))))....(((((((((........)))))))))..... ( -29.00, z-score = -2.05, R) >droEre2.scaffold_4820 9856890 112 + 10470090 ------UGCCAAAUCCAAGCGAAACGAU-CCAAG-UACAACUA-CAAAACCAAAUUGGGCGUAAUAUU--AUGCCUAGGCUGGGUGAAAUAUGAGCUGAGAAAAUUUUAUUUCAGCUUUGUUU ------......(((((.((........-....(-((....))-).........(((((((((....)--))))))))))))))).(((((.(((((((((........)))))))))))))) ( -28.30, z-score = -2.16, R) >droAna3.scaffold_13340 16501643 100 - 23697760 ------------------CCCAACCGAU-CCAAG-UACAAGUA-CGAAGCCAAAUUGGGCGUAAUAUU--AUGCCCAGGCUGGGUGAAAUAUGAGCUGAGAAAAUUUUAUUUCAGCUUUGCUU ------------------(((.......-....(-((....))-)..((((....((((((((....)--))))))))))))))........(((((((((........)))))))))..... ( -31.50, z-score = -3.25, R) >dp4.chr2 4613331 96 - 30794189 -----------------------ACGAUCCCCAG-UACAAGUA-CAGAGCCAAAUUGGGCGUAAUAUU--AUGCCCAGGCUGGGUGAAAUAUGAGCUGAGAAAAUUUUAUUUCAGCUUUGUUU -----------------------....(((((.(-((....))-)..((((....((((((((....)--)))))))))))))).))((((.(((((((((........))))))))))))). ( -33.70, z-score = -4.37, R) >droPer1.super_19 311995 101 - 1869541 ------------------ACGAAACGAUCCCCAG-UACAAGUA-CAGAGCCAAAUUGGGCGUAAUAUU--AUGCCCAGGCUGGGUGAAAUAUGAGCUGAGAAAAUUUUAUUUCAGCUUUGUUU ------------------...(((((((((((.(-((....))-)..((((....((((((((....)--)))))))))))))).))......((((((((........)))))))))))))) ( -34.70, z-score = -4.51, R) >droWil1.scaffold_181130 13512593 103 + 16660200 --------------------AAACAGAUCCAGAGCCAGCCUUGAAGAAGCCAGCGAGUACAUAAAGCCGAAUUGGUUACCAACUUGAAAUAUUAGCUGAGAAAGCUAGAUUUCUGCAUCGCCU --------------------.....(((.(((((.............(((((.((.((.......))))...)))))..............((((((.....))))))..))))).))).... ( -18.70, z-score = 0.04, R) >droVir3.scaffold_13047 12785304 92 - 19223366 ----------------------------GUCCAG-CACCGUUGCCAGCGCCAAAUUGGGCGUAAUAUU--AUGCCCAGCCUGGGUGAAAUAUGAGCUGAGAAAAUUUUAUUUCUGCUUUGCAU ----------------------------.....(-((......((((.((.....((((((((....)--))))))))))))).........((((.((((........)))).))))))).. ( -26.50, z-score = -1.01, R) >droMoj3.scaffold_6540 5092526 92 + 34148556 ----------------------------GUCCAG-CACCGUUGCCAACGCCAAAUUGGGCGUAAUAUU--AUGCCCAGGCUGGGUGAAAUAUGAGCUGAGAAAAUUUUAUUUCUGCUUUGUUU ----------------------------.(((((-(..(((.....))).....(((((((((....)--))))))))))))))..(((((.((((.((((........)))).))))))))) ( -28.20, z-score = -1.96, R) >droGri2.scaffold_15074 5819139 92 + 7742996 ----------------------------GUCCAG-CACCGUUGCCAGCGCCAAAUUGGGCGUAAUAUU--AUGCCCAGCCUGGGUGAAAUAUGAGCUGAGAAAAUUUUAUUUCUGCUUUGUUU ----------------------------...(((-(..(((.....((.(((..(((((((((....)--))))))))..))))).....))).))))(((((......)))))......... ( -25.80, z-score = -1.16, R) >consensus ___________________CGAAACGAU_CCAAG_UACAACUA_CAGAGCCAAAUUGGGCGUAAUAUU__AUGCCCAGGCUGGGUGAAAUAUGAGCUGAGAAAAUUUUAUUUCAGCUUUGUUU .................................................(((..((((((((........))))))))..))).........((((.((((........)))).))))..... (-17.91 = -18.06 + 0.15)

| Location | 27,366,489 – 27,366,607 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 77.47 |
| Shannon entropy | 0.38461 |
| G+C content | 0.48171 |
| Mean single sequence MFE | -26.02 |
| Consensus MFE | -20.08 |
| Energy contribution | -20.12 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.15 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.734802 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
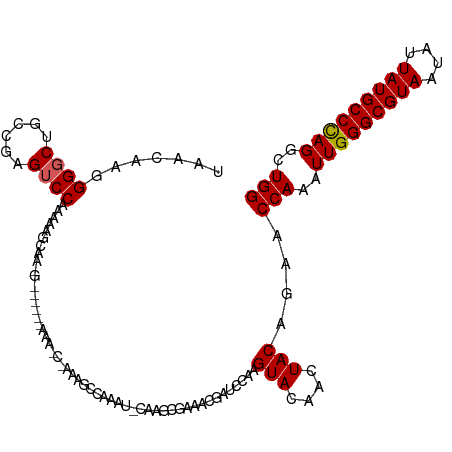
>dm3.chr3R 27366489 118 - 27905053 UAACAAGGGUCCGCCGAGUCCAAAAAGCAACGAAAACCAAAGCCAAAGCCAAAUCCAAGCGAAACGAUCCAUGUACAACUACAGAACCAAAUUGGGCGUAAUAUUAUGCCCAGGCUGG ..(((.((((((((...((.......)).............((....)).........)))....))))).)))............(((..(((((((((....)))))))))..))) ( -25.40, z-score = -1.22, R) >droYak2.chr3R 28301201 112 - 28832112 UAACAAGGGGCUGCCGAGUCCAAAAAGCAACG------AAAACCAAUGCCAAAUUCAAGCGAAACGAUCCGAGUACAACUACAGAACCAAAUUGGGCGUAAUAUUAUGCCCAGGCUGG .......(((((....)))))...........------....(((..(((...(((...((........)).(((....))).)))......((((((((....)))))))))))))) ( -24.70, z-score = -0.62, R) >droEre2.scaffold_4820 9856930 112 + 10470090 UAACAAGGGGCUGCCGAGUCCAAAAAGCAAGG------AAAGCUAAUGCCAAAUCCAAGCGAAACGAUCCAAGUACAACUACAAAACCAAAUUGGGCGUAAUAUUAUGCCUAGGCUGG .......(((((....))))).....((..((------(..((....))....)))..))..........................(((..(((((((((....)))))))))..))) ( -25.70, z-score = -0.50, R) >droAna3.scaffold_13340 16501683 100 - 23697760 UAACAAGGGGCUCCCGAGUCCGAAAGGC----------------AACGAAAAG--CAACCCAACCGAUCCAAGUACAAGUACGAAGCCAAAUUGGGCGUAAUAUUAUGCCCAGGCUGG ......((((((..((...((....)).----------------..))...))--)..)))..((.......(((....)))..((((....((((((((....)))))))))))))) ( -29.80, z-score = -2.07, R) >dp4.chr2 4613371 92 - 30794189 UAACAAGGGGCCCCCGAGUCCGAGAAAC----------------GAAACGAAA--CGAUC--------CCCAGUACAAGUACAGAGCCAAAUUGGGCGUAAUAUUAUGCCCAGGCUGG ......((((....((..((((.....)----------------)....))..--))..)--------))).(((....)))..((((....((((((((....)))))))))))).. ( -24.50, z-score = -1.35, R) >consensus UAACAAGGGGCUGCCGAGUCCAAAAAGCAA_G______AAA_C_AAAGCCAAAU_CAAGCGAAACGAUCCAAGUACAACUACAGAACCAAAUUGGGCGUAAUAUUAUGCCCAGGCUGG .......((((......))))...................................................(((....)))....(((..(((((((((....)))))))))..))) (-20.08 = -20.12 + 0.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:59:59 2011