| Sequence ID | dm3.chr3R |
|---|---|
| Location | 27,346,833 – 27,346,923 |
| Length | 90 |
| Max. P | 0.950514 |

| Location | 27,346,833 – 27,346,923 |
|---|---|
| Length | 90 |
| Sequences | 9 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 68.25 |
| Shannon entropy | 0.62841 |
| G+C content | 0.55557 |
| Mean single sequence MFE | -29.91 |
| Consensus MFE | -14.83 |
| Energy contribution | -15.33 |
| Covariance contribution | 0.51 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.56 |
| SVM RNA-class probability | 0.950514 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
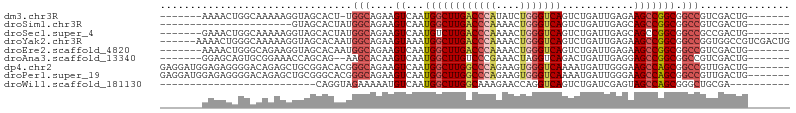
>dm3.chr3R 27346833 90 - 27905053 -------AAAACUGGCAAAAAGGUAGCACU-UGGCAGAAGUCAAUGGCUUGACCCAUAUCUGGGUCAGUCUGAUUGAGAAGCCGGCGGCCGUCGACUG------- -------....(((.((...((......))-)).))).((((.(((((((((((((....)))))))(((.(.((....))).))))))))).)))).------- ( -29.10, z-score = -1.16, R) >droSim1.chr3R 26982427 76 - 27517382 ----------------------GUAGCACUAUGGCAGAAGUCAAUGGCUUGACCCAAAACUGGGUCAGUCUGAUUGAGCAGCCGGCGGCCGUCGACUG------- ----------------------...((......))...((((.(((((((((((((....)))))))(.(((......))).)...)))))).)))).------- ( -26.70, z-score = -1.31, R) >droSec1.super_4 6146644 91 - 6179234 -------GAAACUGGCAAAAAGGUAGCACUAUGGCAGAAGUCAAUGUCUUGACCCAAAACUGGGUCAGUCUGAUUGAGCAGCCGGCGGCCGCCGACUG------- -------....(((.((...((......)).)).)))...(((((...((((((((....))))))))....))))).(((.((((....)))).)))------- ( -28.30, z-score = -0.78, R) >droYak2.chr3R 28281258 99 - 28832112 ------AAAACUGGGCAAAAAGGUAGCACAAUGGCAGAAGUAAAUGGCUUGACCCAAAACUGGGUCAGUCUGAUUGAGAAGCCGGCGGCCGGUGGCCGUCGACUG ------.......(((.........((......))..........((((.((((((....))))))))))..........)))(((((((...)))))))..... ( -33.50, z-score = -1.41, R) >droEre2.scaffold_4820 9837378 91 + 10470090 -------AAAACUGGGCAGAAGGUAGCACAAUGGCAGAAGUCAAUGGCUUGACCCAAAACUGGGUCAGUCUGAUUGAGAAGCCGGCGGCCGUCGACUG------- -------.........(((..((..((....((((...(((((..((((.((((((....))))))))))))))).....))))...))..))..)))------- ( -29.80, z-score = -1.16, R) >droAna3.scaffold_13340 16478703 89 - 23697760 -------GGAGCAGUGCGGAAACCAGCAG--AAGCACAAGUCAAUGGCUUGUCCCGAAACUAGGUCAGACUGAUUGAGGAGCCGGCGGCCGUCGACUG------- -------......(((((....)(....)--..)))).((((.((((((.(.((.(...((..(((.....)))..))...).))))))))).)))).------- ( -24.50, z-score = 0.78, R) >dp4.chr2 4587505 98 - 30794189 GAGGAUGGAGAGGGGACAGAGCUGCGGACACGGGCAGAAGUCAAUGGCUUGGCCCAGAAGUGGGUCAAAAUGAUUGGGAAGCCAGCGGCCGUUGACUG------- ...........(....)....((((........)))).((((((((((((((((((....))))))))..((.((((....)))))))))))))))).------- ( -37.60, z-score = -3.17, R) >droPer1.super_19 1620205 98 + 1869541 GAGGAUGGAGAGGGGACAGAGCUGCGGGCACGGGCAGAAGUCAAUGGCUUGGCCCAGAAGUGGGUCAAAAUGAUUGGGAAGCCAGCGGCCGUUGACUG------- ...........(....)....((((........)))).((((((((((((((((((....))))))))..((.((((....)))))))))))))))).------- ( -37.60, z-score = -2.73, R) >droWil1.scaffold_181130 13486273 69 + 16660200 --------------------------CAGGUAGAAAAAUGUCAAUGGCUUGGCAAAGAACCAGGUCAGUCUGAUCGAGUAGCCAGCGGGCUGCGA---------- --------------------------.............((((.((((((((.......))))))))...))))...((((((....))))))..---------- ( -22.10, z-score = -1.88, R) >consensus _______GAAACGGGCCAGAAGGUAGCACAAUGGCAGAAGUCAAUGGCUUGACCCAAAACUGGGUCAGUCUGAUUGAGAAGCCGGCGGCCGUCGACUG_______ ................................(((....((...((((((((((((....)))))))............))))))).)))............... (-14.83 = -15.33 + 0.51)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:59:55 2011