
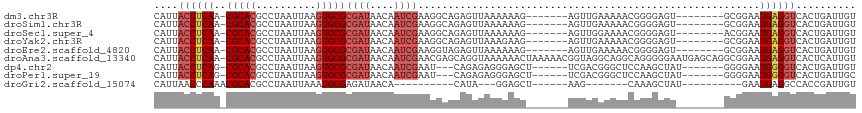
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 27,338,927 – 27,339,028 |
| Length | 101 |
| Max. P | 0.693405 |

| Location | 27,338,927 – 27,339,028 |
|---|---|
| Length | 101 |
| Sequences | 9 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 75.02 |
| Shannon entropy | 0.47897 |
| G+C content | 0.46584 |
| Mean single sequence MFE | -26.43 |
| Consensus MFE | -12.85 |
| Energy contribution | -13.59 |
| Covariance contribution | 0.74 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.672485 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 27338927 101 + 27905053 CAUUACCUCAA-CGCACGCCUAAUUAAGUGCGCGAUAACAAUCGAAGGCAGAGUUAAAAAAG-------AGUUGAAAAACGGGGAGU--------GCGGAAUGAGGUCACUGAUUGU ....((((((.-(((((.(((.......(((.((((....))))...)))............-------.(((....))).))).))--------)))...)))))).......... ( -28.70, z-score = -2.63, R) >droSim1.chr3R 26968541 101 + 27517382 CAUUACCUCAA-CGCACGCCUAAUUAAGUGCGCGAUAACAAUCGAAGGCAGAGUUAAAAAAG-------AGUUGAAAAACGGGGAGU--------GCGGAAUGAGGUCACUGAUUGU ....((((((.-(((((.(((.......(((.((((....))))...)))............-------.(((....))).))).))--------)))...)))))).......... ( -28.70, z-score = -2.63, R) >droSec1.super_4 6138824 101 + 6179234 CAUUACCUCAA-CGCACGCCUAAUUAAGUGCGCGAUAACAAUCGAAGGCAGAGUUAAAAAAG-------AGUUGGAAAACGGGGAGU--------ACGGAAUGAGGUCACUGAUUGU ....((((((.-((.((.(((.......(((.((((....))))...)))............-------.(((....))).))).))--------.))...)))))).......... ( -22.10, z-score = -0.62, R) >droYak2.chr3R 28273165 101 + 28832112 CAUUACCUCAA-CGCACGCCUAAUUAAGUGCGCGAUAACAAUCGAAGGCAGAGUUAAAGAAG-------AGUUGAAAAACGGGGAGU--------GCGGAAUGAGGUCACUGAUUGU ....((((((.-(((((.(((.......(((.((((....))))...)))............-------.(((....))).))).))--------)))...)))))).......... ( -28.70, z-score = -2.49, R) >droEre2.scaffold_4820 9829405 101 - 10470090 CAUUACCUCAA-CGCACGCCUAAUUAAGUGCGCGAUAACAAUCGAAGGUAGAGUUAAAAAAG-------AGUUGAAAAACGGGGAGU--------GCGGAAUGAGGUCACUGAUUGU ....((((((.-(((((.(((.......(((.((((....))))...)))............-------.(((....))).))).))--------)))...)))))).......... ( -26.70, z-score = -2.17, R) >droAna3.scaffold_13340 16470239 116 + 23697760 CAUUACCUCAA-CGCACGCCUAAUUAAGUGCGCGAUAACAAUCGAACGAGCAGGUAAAAAACUAAAAACGGUAGGCAGGCAGGGGGAAUGAGCAGGCGGAAUGAGGUCACUCAUUGU ((((.((((..-.((..((((..(((..(((.((((....)))).....)))..)))...(((......)))))))..))..)))))))).........((((((....)))))).. ( -29.10, z-score = -1.30, R) >dp4.chr2 4577574 100 + 30794189 CAUUACCUCAG-CGCACGCCUAAUUAAGUGCGCGAUAACAAUCGAAU---CAGAGAGGGAGCU------UCGACGGGCUCCAAGCUAU-------GGGGAAUGGGGUCACUGAUUGU ((((.((((((-(((((..........))))))..............---...((..((((((------......))))))...)).)-------)))))))).............. ( -31.20, z-score = -1.32, R) >droPer1.super_19 1610175 100 - 1869541 CAUUACCUCAG-CGCACGCCUAAUUAAGUGCGCGAUAACAAUCGAAU---CAGAGAGGGAGCU------UCGACGGGCUCCAAGCUAU-------GGGGAAUGGGGUCACUGAUUGC .....((((.(-(((((..........))))))(((....)))....---....))))..((.------(((...(((((((..((..-------..))..)))))))..)))..)) ( -31.30, z-score = -1.38, R) >droGri2.scaffold_15074 5785575 81 - 7742996 CAUUAACCCAAACGCACGCCUAAUUAAAUGCGAGAUAACA----------CAUA---GGAGCU------AAG-------CAAAGCUAU----------GAAUGAGGCCACCGAUUGU ........(((.((...((((.(((..(((.(......).----------))).---..((((------...-------...))))..----------.))).))))...)).))). ( -11.40, z-score = 0.10, R) >consensus CAUUACCUCAA_CGCACGCCUAAUUAAGUGCGCGAUAACAAUCGAAGGCAGAGUUAAAAAAG_______AGUUGAAAAACGGGGAGU________GCGGAAUGAGGUCACUGAUUGU ....((((((..(((((..........)))))((((....)))).........................................................)))))).......... (-12.85 = -13.59 + 0.74)


| Location | 27,338,927 – 27,339,028 |
|---|---|
| Length | 101 |
| Sequences | 9 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 75.02 |
| Shannon entropy | 0.47897 |
| G+C content | 0.46584 |
| Mean single sequence MFE | -22.03 |
| Consensus MFE | -12.40 |
| Energy contribution | -12.91 |
| Covariance contribution | 0.51 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.693405 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 27338927 101 - 27905053 ACAAUCAGUGACCUCAUUCCGC--------ACUCCCCGUUUUUCAACU-------CUUUUUUAACUCUGCCUUCGAUUGUUAUCGCGCACUUAAUUAGGCGUGCG-UUGAGGUAAUG .....((...((((((...(((--------((.((..(((....))).-------............(((...((((....)))).)))........)).)))))-.))))))..)) ( -23.00, z-score = -2.41, R) >droSim1.chr3R 26968541 101 - 27517382 ACAAUCAGUGACCUCAUUCCGC--------ACUCCCCGUUUUUCAACU-------CUUUUUUAACUCUGCCUUCGAUUGUUAUCGCGCACUUAAUUAGGCGUGCG-UUGAGGUAAUG .....((...((((((...(((--------((.((..(((....))).-------............(((...((((....)))).)))........)).)))))-.))))))..)) ( -23.00, z-score = -2.41, R) >droSec1.super_4 6138824 101 - 6179234 ACAAUCAGUGACCUCAUUCCGU--------ACUCCCCGUUUUCCAACU-------CUUUUUUAACUCUGCCUUCGAUUGUUAUCGCGCACUUAAUUAGGCGUGCG-UUGAGGUAAUG .....((...((((((...(((--------((.((..(((....))).-------............(((...((((....)))).)))........)).)))))-.))))))..)) ( -21.00, z-score = -1.87, R) >droYak2.chr3R 28273165 101 - 28832112 ACAAUCAGUGACCUCAUUCCGC--------ACUCCCCGUUUUUCAACU-------CUUCUUUAACUCUGCCUUCGAUUGUUAUCGCGCACUUAAUUAGGCGUGCG-UUGAGGUAAUG .....((...((((((...(((--------((.((..(((....))).-------............(((...((((....)))).)))........)).)))))-.))))))..)) ( -23.00, z-score = -2.40, R) >droEre2.scaffold_4820 9829405 101 + 10470090 ACAAUCAGUGACCUCAUUCCGC--------ACUCCCCGUUUUUCAACU-------CUUUUUUAACUCUACCUUCGAUUGUUAUCGCGCACUUAAUUAGGCGUGCG-UUGAGGUAAUG .....((...((((((...(((--------((................-------..................((((....)))).((.((.....)))))))))-.))))))..)) ( -20.90, z-score = -2.12, R) >droAna3.scaffold_13340 16470239 116 - 23697760 ACAAUGAGUGACCUCAUUCCGCCUGCUCAUUCCCCCUGCCUGCCUACCGUUUUUAGUUUUUUACCUGCUCGUUCGAUUGUUAUCGCGCACUUAAUUAGGCGUGCG-UUGAGGUAAUG ..........((((((...((((.((...........))..(((((........(((.........))).((.((((....)))).)).......)))))).)))-.)))))).... ( -22.50, z-score = -0.25, R) >dp4.chr2 4577574 100 - 30794189 ACAAUCAGUGACCCCAUUCCCC-------AUAGCUUGGAGCCCGUCGA------AGCUCCCUCUCUG---AUUCGAUUGUUAUCGCGCACUUAAUUAGGCGUGCG-CUGAGGUAAUG ((((((((((....))))...(-------(.((...(((((.......------.)))))..)).))---....))))))..((((((((..........)))))-).))....... ( -23.80, z-score = -0.30, R) >droPer1.super_19 1610175 100 + 1869541 GCAAUCAGUGACCCCAUUCCCC-------AUAGCUUGGAGCCCGUCGA------AGCUCCCUCUCUG---AUUCGAUUGUUAUCGCGCACUUAAUUAGGCGUGCG-CUGAGGUAAUG ((((((((((....))))...(-------(.((...(((((.......------.)))))..)).))---....))))))..((((((((..........)))))-).))....... ( -24.10, z-score = -0.11, R) >droGri2.scaffold_15074 5785575 81 + 7742996 ACAAUCGGUGGCCUCAUUC----------AUAGCUUUG-------CUU------AGCUCC---UAUG----------UGUUAUCUCGCAUUUAAUUAGGCGUGCGUUUGGGUUAAUG ........(((((.((...----------...((..((-------(((------((....---.(((----------((......)))))....))))))).))...)))))))... ( -17.00, z-score = -0.11, R) >consensus ACAAUCAGUGACCUCAUUCCGC________ACUCCCCGUUUUCCAACU_______CUUUUUUAACUCUGCCUUCGAUUGUUAUCGCGCACUUAAUUAGGCGUGCG_UUGAGGUAAUG ..........((((((..........................................................(((....)))((((.(((....))).))))...)))))).... (-12.40 = -12.91 + 0.51)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:59:55 2011