
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 9,363,968 – 9,364,060 |
| Length | 92 |
| Max. P | 0.810843 |

| Location | 9,363,968 – 9,364,060 |
|---|---|
| Length | 92 |
| Sequences | 8 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 59.13 |
| Shannon entropy | 0.74497 |
| G+C content | 0.39108 |
| Mean single sequence MFE | -20.52 |
| Consensus MFE | -7.80 |
| Energy contribution | -7.41 |
| Covariance contribution | -0.39 |
| Combinations/Pair | 1.64 |
| Mean z-score | -1.08 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.734031 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
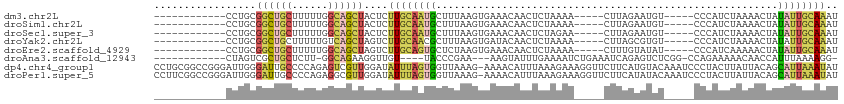
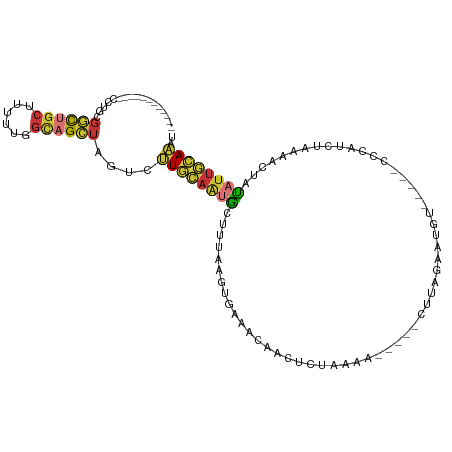
>dm3.chr2L 9363968 92 + 23011544 ------------CCUGCGGCUGCUUUUUGGCAGCUACUCUUGCAAUGCUUUAAGUGAAACAACUCUAAAA-----CUUAGAAUGU-----CCCAUCUAAAACUAUAUUGCAAAU ------------...(..((((((....))))))..)..((((((((.((((..((..(((..((((...-----..)))).)))-----..))..))))....)))))))).. ( -23.00, z-score = -3.28, R) >droSim1.chr2L 9141322 92 + 22036055 ------------CCUGCGGCUGCUUUUUGGCAGCUACUCUUGCAAUGCUUUAAGUGAAACAACUCUAAAA-----CUUAGAAUGU-----CCCAUCUAAAACUAUAUUGCAAAU ------------...(..((((((....))))))..)..((((((((.((((..((..(((..((((...-----..)))).)))-----..))..))))....)))))))).. ( -23.00, z-score = -3.28, R) >droSec1.super_3 4821410 92 + 7220098 ------------CCUGCGGCUGCUUUUUGGCAGCUACUCUUGCAAUGCUUUAAGUGAAACAACUCUAGAA-----CUUAGAAUGU-----CCCAUCUAAAACUAUAUUGCAAAU ------------...(..((((((....))))))..)..((((((((.((((..((..(((..(((((..-----.))))).)))-----..))..))))....)))))))).. ( -22.70, z-score = -2.51, R) >droYak2.chr2L 12036430 92 + 22324452 ------------CCUGCGGCUGCUUUUUGUCAGCUAGUCUUGCAACGCUUUAAGUGAUACAACUCUAAAA-----CUUAGCGUGU-----CCCAUCUAAAACUAUAUUGCAAAU ------------..(((((((((.....).))))(((...((..((((.((((((..............)-----))))).))))-----..)).))).........))))... ( -17.14, z-score = -0.51, R) >droEre2.scaffold_4929 9973996 92 + 26641161 ------------CCUGCGGCUGCUUUUUGGCAGCUAGUCUUGCAGUGCUCUAAGUGAAACAACUCUAAAA-----CUUUGUAUAU-----CCCAUCAAAAACUAUAUUGCAAAU ------------.....(((((((....)))))))....((((((((.....(((......)))......-----.((((.....-----.....)))).....)))))))).. ( -18.00, z-score = -0.61, R) >droAna3.scaffold_12943 716965 92 + 5039921 ------------CUAGUCGCUGCUCUU-GGCAGAAGGUUGU----UACCCGAA---AAGUAUUUGAAAAUCUGAAAUCAGAGUCUCGG-CCAGAAAAACAACCAUUUAAAAGG- ------------.......((((....-.))))..((((((----(..((((.---........((...((((....)))).))))))-.......)))))))..........- ( -19.40, z-score = -0.66, R) >dp4.chr4_group1 4337537 113 - 5278887 CCUGCGGCCGGGAUUGGGAUUGCCCCAGAGUCGUUGGAUAUUUAGUGGUUAAAG-AAAACAUUUAAAGAAAGGUUCUUCAUGUACAAAUCCCUACUUAUUACAGCAUUAAAUAU ((.(((((.....(((((.....))))).))))).))((((((((((......(-(((((.(((....))).))).))).((((...............)))).)))))))))) ( -20.76, z-score = 1.09, R) >droPer1.super_5 4267380 113 + 6813705 CCUUCGGCCGGGAUUGGGAUUGCCCCAGAGGCGUUGGAUAUUUAGUGGUUAAAG-AAAACAUUUAAAGAAAGGUUCUUCAUAUACAAAUCCCUACUUAUUACAGCAUUAAAUAU ......(((....(((((.....))))).)))((((((((..(((.(((((((.-......))))).(((......)))..........)))))..)))).))))......... ( -20.20, z-score = 1.13, R) >consensus ____________CCUGCGGCUGCUUUUUGGCAGCUAGUCUUGCAAUGCUUUAAGUGAAACAACUCUAAAA_____CUUAGAAUGU_____CCCAUCUAAAACUAUAUUGCAAAU .................((((((......))))))....((((((((.........................................................)))))))).. ( -7.80 = -7.41 + -0.39)
| Location | 9,363,968 – 9,364,060 |
|---|---|
| Length | 92 |
| Sequences | 8 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 59.13 |
| Shannon entropy | 0.74497 |
| G+C content | 0.39108 |
| Mean single sequence MFE | -24.96 |
| Consensus MFE | -6.84 |
| Energy contribution | -6.19 |
| Covariance contribution | -0.65 |
| Combinations/Pair | 1.82 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.27 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.810843 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
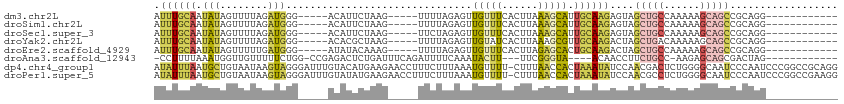
>dm3.chr2L 9363968 92 - 23011544 AUUUGCAAUAUAGUUUUAGAUGGG-----ACAUUCUAAG-----UUUUAGAGUUGUUUCACUUAAAGCAUUGCAAGAGUAGCUGCCAAAAAGCAGCCGCAGG------------ .((((((((...(((((((.((..-----((((((((..-----...))))).)))..)).))))))))))))))).(..(((((......)))))..)...------------ ( -32.10, z-score = -4.71, R) >droSim1.chr2L 9141322 92 - 22036055 AUUUGCAAUAUAGUUUUAGAUGGG-----ACAUUCUAAG-----UUUUAGAGUUGUUUCACUUAAAGCAUUGCAAGAGUAGCUGCCAAAAAGCAGCCGCAGG------------ .((((((((...(((((((.((..-----((((((((..-----...))))).)))..)).))))))))))))))).(..(((((......)))))..)...------------ ( -32.10, z-score = -4.71, R) >droSec1.super_3 4821410 92 - 7220098 AUUUGCAAUAUAGUUUUAGAUGGG-----ACAUUCUAAG-----UUCUAGAGUUGUUUCACUUAAAGCAUUGCAAGAGUAGCUGCCAAAAAGCAGCCGCAGG------------ .((((((((...(((((((.((..-----((((((((..-----...))))).)))..)).))))))))))))))).(..(((((......)))))..)...------------ ( -32.30, z-score = -4.46, R) >droYak2.chr2L 12036430 92 - 22324452 AUUUGCAAUAUAGUUUUAGAUGGG-----ACACGCUAAG-----UUUUAGAGUUGUAUCACUUAAAGCGUUGCAAGACUAGCUGACAAAAAGCAGCCGCAGG------------ .((((((((...(((((((.((..-----((((.(((..-----...))).).)))..)).)))))))))))))))....((((........))))......------------ ( -20.60, z-score = -0.30, R) >droEre2.scaffold_4929 9973996 92 - 26641161 AUUUGCAAUAUAGUUUUUGAUGGG-----AUAUACAAAG-----UUUUAGAGUUGUUUCACUUAGAGCACUGCAAGACUAGCUGCCAAAAAGCAGCCGCAGG------------ ..((((....(((((((...((..-----.....))..(-----((((.((((......))))))))).....)))))))(((((......))))).)))).------------ ( -19.50, z-score = 0.06, R) >droAna3.scaffold_12943 716965 92 - 5039921 -CCUUUUAAAUGGUUGUUUUUCUGG-CCGAGACUCUGAUUUCAGAUUUUCAAAUACUU---UUCGGGUA----ACAACCUUCUGCC-AAGAGCAGCGACUAG------------ -.........((((((((.((((((-(.((((.((((....)))).))))........---...((((.----...))))...)))-.)))).)))))))).------------ ( -26.40, z-score = -2.96, R) >dp4.chr4_group1 4337537 113 + 5278887 AUAUUUAAUGCUGUAAUAAGUAGGGAUUUGUACAUGAAGAACCUUUCUUUAAAUGUUUU-CUUUAACCACUAAAUAUCCAACGACUCUGGGGCAAUCCCAAUCCCGGCCGCAGG (((((((.((((......))))((.....(.(((((((((.....)))))..))))...-).....))..))))))).........(((.(((.............))).))). ( -18.32, z-score = 1.06, R) >droPer1.super_5 4267380 113 - 6813705 AUAUUUAAUGCUGUAAUAAGUAGGGAUUUGUAUAUGAAGAACCUUUCUUUAAAUGUUUU-CUUUAACCACUAAAUAUCCAACGCCUCUGGGGCAAUCCCAAUCCCGGCCGAAGG ........((((......))))((((((..(((.((((((.....)))))).)))....-......................(((.....)))))))))............... ( -18.40, z-score = 1.07, R) >consensus AUUUGCAAUAUAGUUUUAGAUGGG_____ACAUUCUAAG_____UUUUAGAGUUGUUUCACUUAAAGCAUUGCAAGACUAGCUGCCAAAAAGCAGCCGCAGG____________ .((((((.........((((((........))).)))................(((((......))))).))))))....(((((......))))).................. ( -6.84 = -6.19 + -0.65)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:28:02 2011