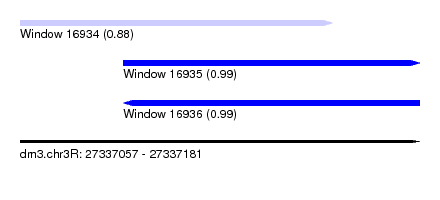
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 27,337,057 – 27,337,181 |
| Length | 124 |
| Max. P | 0.992960 |

| Location | 27,337,057 – 27,337,154 |
|---|---|
| Length | 97 |
| Sequences | 10 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 78.12 |
| Shannon entropy | 0.43600 |
| G+C content | 0.47787 |
| Mean single sequence MFE | -21.59 |
| Consensus MFE | -13.14 |
| Energy contribution | -13.35 |
| Covariance contribution | 0.21 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.881242 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
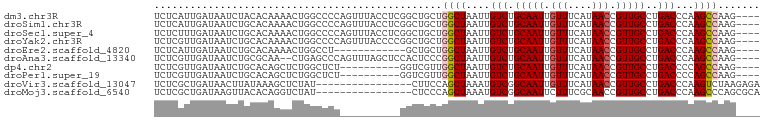
>dm3.chr3R 27337057 97 + 27905053 UCUCAUUGAUAAUCUACACAAAACUGGCCCCAGUUUACCUCGGCUGCUGGCUAAUUGUCUGCAAUUGUUUCAUAACCGUUGCCUGACCCAAGCCAAG---- ....................((((((....)))))).....((((..(((......(((.(((((.(((....))).)))))..))))))))))...---- ( -22.30, z-score = -1.88, R) >droSim1.chr3R 26966660 97 + 27517382 UCUCAUUGAUAAUCUGCACAAAACUGGCCCCAGUUUACCUCGGCUGCUGGCUAAUUGUCUGCAAUUGUUUCAUAACCGUUGCCUGACCCAAGCCAAG---- ...............((...((((((....))))))......))...(((((....(((.(((((.(((....))).)))))..)))...)))))..---- ( -22.80, z-score = -1.58, R) >droSec1.super_4 6136940 97 + 6179234 UCUCUUUGAUAAUCUGCACAAAACUGGCCCCAGUUUACCUCGGCUGCUGGCUAAUUGUCUGCAAUUGUUUCAUAACCGUUGCCUGACCCAAGCCAAG---- ...............((...((((((....))))))......))...(((((....(((.(((((.(((....))).)))))..)))...)))))..---- ( -22.80, z-score = -1.71, R) >droYak2.chr3R 28271213 97 + 28832112 UCUCGUUGAUAAUCUGCACAAAACUGGCCCCAGUUUACCCCGGCUGCUGGCUAAUUGUCUGCAAUUGUUUCAUAACCGUUGCCUGACCCAAGCCAAG---- ...............((...((((((....))))))......))...(((((....(((.(((((.(((....))).)))))..)))...)))))..---- ( -22.80, z-score = -1.42, R) >droEre2.scaffold_4820 9827514 85 - 10470090 UCUCAUUGAUAAUCUGCACAAAACUGGCCU------------GCUGCUGGCUAAUUGUCUGCAAUUGUUUCAUAACCGUUGCCUGACCCAAGCCAAG---- ...............(((((.........)------------).)))(((((....(((.(((((.(((....))).)))))..)))...)))))..---- ( -17.50, z-score = -0.68, R) >droAna3.scaffold_13340 16468147 95 + 23697760 UCUCGUUGAUAAUCUGCGCAA--CUGAGCCCAGUUUAGCUCCACUCCCGGCUAAUUGUCUGCAAUUGUUUCAUAACCGUUGCCUGACCCAAGCCAAG---- ....(((((....(((.((..--....)).))).))))).........((((....(((.(((((.(((....))).)))))..)))...))))...---- ( -23.20, z-score = -2.51, R) >dp4.chr2 26231711 87 - 30794189 UCUCGUUGAUAAUCUGCACAGCUCUGGCUCU----------GGUCGUUGGCUAAUUGUCUGCAAUUGUUUCAUAACCGUUGCCUGACCCCAGCCAAG---- ....((((..........))))..(((((..----------((((...(((.....))).(((((.(((....))).)))))..))))..)))))..---- ( -22.90, z-score = -1.67, R) >droPer1.super_19 274489 87 + 1869541 UCUCGUUGAUAAUCUGCACAGCUCUGGCUCU----------GGUCGUUGGCUAAUUGUCUGCAAUUGUUUCAUAACCGUUGCCUGACCCCAGCCAAG---- ....((((..........))))..(((((..----------((((...(((.....))).(((((.(((....))).)))))..))))..)))))..---- ( -22.90, z-score = -1.67, R) >droVir3.scaffold_13047 12750218 85 + 19223366 UCUCGCUGAUAACUUAUAAAGCUCUAU----------------CUUCCAGCUAAAUGUCGGCAAUUGUUUCAUAACCGUUGCCUGACCCAAGUCUAAGAGA ((((...(((.........((((....----------------.....))))....(((((((((.(((....))).)))))).)))....)))...)))) ( -20.00, z-score = -2.70, R) >droMoj3.scaffold_6540 5059862 85 - 34148556 UCUCGCUGAUAAGUUACACAGGUCUAU----------------CUCCCAGCUAAAUGUCGGCAAUUCUUUCGCAACCGUUGCCUGACCCAAGCCCAGCGCA ...(((((............((.....----------------...)).(((....(((((((((............)))))).)))...))).))))).. ( -18.70, z-score = -0.94, R) >consensus UCUCGUUGAUAAUCUGCACAAAACUGGCCCC__________GGCUGCUGGCUAAUUGUCUGCAAUUGUUUCAUAACCGUUGCCUGACCCAAGCCAAG____ ................................................((((....(((.(((((.(((....))).)))))..)))...))))....... (-13.14 = -13.35 + 0.21)
| Location | 27,337,089 – 27,337,181 |
|---|---|
| Length | 92 |
| Sequences | 8 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 87.70 |
| Shannon entropy | 0.22872 |
| G+C content | 0.51989 |
| Mean single sequence MFE | -25.71 |
| Consensus MFE | -19.45 |
| Energy contribution | -20.07 |
| Covariance contribution | 0.63 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.14 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.58 |
| SVM RNA-class probability | 0.992960 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 27337089 92 + 27905053 GUUUACCUCGGCUGCUGGCUAAUUGUCUGCAAUUGUUUCAUAACCGUUGCCUGACCCAAGCCAAGUGACCCAAAUCCGCUG-------ACCAUCCAGCC .........((..(((((((....(((.(((((.(((....))).)))))..)))...)))).)))..)).......((((-------......)))). ( -25.50, z-score = -2.97, R) >droSim1.chr3R 26966692 92 + 27517382 GUUUACCUCGGCUGCUGGCUAAUUGUCUGCAAUUGUUUCAUAACCGUUGCCUGACCCAAGCCAAGUGACCCAAAUCCGCUG-------ACCAUCCAGCC .........((..(((((((....(((.(((((.(((....))).)))))..)))...)))).)))..)).......((((-------......)))). ( -25.50, z-score = -2.97, R) >droSec1.super_4 6136972 92 + 6179234 GUUUACCUCGGCUGCUGGCUAAUUGUCUGCAAUUGUUUCAUAACCGUUGCCUGACCCAAGCCAAGUGACCCAAAUCCGCUG-------ACCAUCCAGCC .........((..(((((((....(((.(((((.(((....))).)))))..)))...)))).)))..)).......((((-------......)))). ( -25.50, z-score = -2.97, R) >droYak2.chr3R 28271245 92 + 28832112 GUUUACCCCGGCUGCUGGCUAAUUGUCUGCAAUUGUUUCAUAACCGUUGCCUGACCCAAGCCAAGUGACCCAAAUCCGCUG-------ACCAUCCAGCC .........((..(((((((....(((.(((((.(((....))).)))))..)))...)))).)))..)).......((((-------......)))). ( -25.50, z-score = -3.13, R) >droEre2.scaffold_4820 9827544 82 - 10470090 ----------GCUGCUGGCUAAUUGUCUGCAAUUGUUUCAUAACCGUUGCCUGACCCAAGCCAAGUGACCCAAAUCCACUG-------ACCAUCCAGCC ----------((((.(((((....(((.(((((.(((....))).)))))..)))...)))))((((.........)))).-------......)))). ( -22.70, z-score = -3.43, R) >droAna3.scaffold_13340 16468177 99 + 23697760 GUUUAGCUCCACUCCCGGCUAAUUGUCUGCAAUUGUUUCAUAACCGUUGCCUGACCCAAGCCAAGUGACCCAAAUCUGCUGGCGGCUAACCAUCCAGCC .........((((...((((....(((.(((((.(((....))).)))))..)))...)))).))))..........(((((.((....))..))))). ( -26.40, z-score = -2.26, R) >dp4.chr2 26231741 84 - 30794189 --------UGGUCGUUGGCUAAUUGUCUGCAAUUGUUUCAUAACCGUUGCCUGACCCCAGCCAAGUGACCCAAAUCUUCUG-------GCCAUCCAGCC --------.(((((((((((....(((.(((((.(((....))).)))))..)))...)))))).)))))........(((-------(....)))).. ( -27.30, z-score = -3.72, R) >droPer1.super_19 274519 84 + 1869541 --------UGGUCGUUGGCUAAUUGUCUGCAAUUGUUUCAUAACCGUUGCCUGACCCCAGCCAAGUGACCCAAAUCUUCUG-------GCCAUCCAGCC --------.(((((((((((....(((.(((((.(((....))).)))))..)))...)))))).)))))........(((-------(....)))).. ( -27.30, z-score = -3.72, R) >consensus GUUUACCUCGGCUGCUGGCUAAUUGUCUGCAAUUGUUUCAUAACCGUUGCCUGACCCAAGCCAAGUGACCCAAAUCCGCUG_______ACCAUCCAGCC .........(((((.(((((....(((.(((((.(((....))).)))))..)))...)))))((((.........))))..............))))) (-19.45 = -20.07 + 0.63)

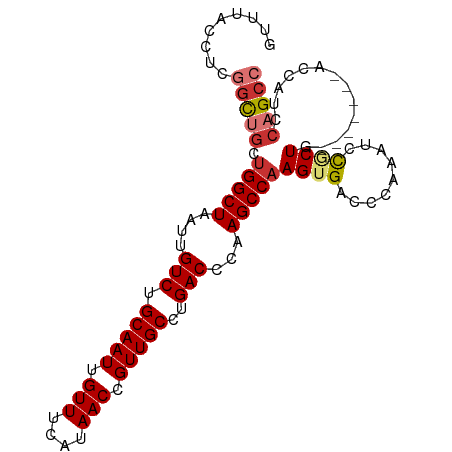
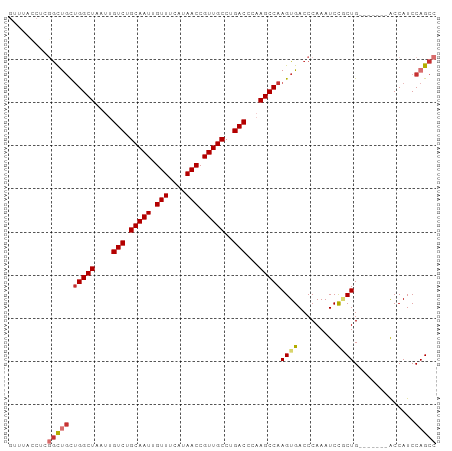
| Location | 27,337,089 – 27,337,181 |
|---|---|
| Length | 92 |
| Sequences | 8 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 87.70 |
| Shannon entropy | 0.22872 |
| G+C content | 0.51989 |
| Mean single sequence MFE | -31.04 |
| Consensus MFE | -26.25 |
| Energy contribution | -26.29 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.85 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.19 |
| SVM RNA-class probability | 0.985139 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 27337089 92 - 27905053 GGCUGGAUGGU-------CAGCGGAUUUGGGUCACUUGGCUUGGGUCAGGCAACGGUUAUGAAACAAUUGCAGACAAUUAGCCAGCAGCCGAGGUAAAC (((((..((((-------.....((((..((((....))))..))))..((((..(((....)))..)))).........)))).)))))......... ( -30.70, z-score = -2.23, R) >droSim1.chr3R 26966692 92 - 27517382 GGCUGGAUGGU-------CAGCGGAUUUGGGUCACUUGGCUUGGGUCAGGCAACGGUUAUGAAACAAUUGCAGACAAUUAGCCAGCAGCCGAGGUAAAC (((((..((((-------.....((((..((((....))))..))))..((((..(((....)))..)))).........)))).)))))......... ( -30.70, z-score = -2.23, R) >droSec1.super_4 6136972 92 - 6179234 GGCUGGAUGGU-------CAGCGGAUUUGGGUCACUUGGCUUGGGUCAGGCAACGGUUAUGAAACAAUUGCAGACAAUUAGCCAGCAGCCGAGGUAAAC (((((..((((-------.....((((..((((....))))..))))..((((..(((....)))..)))).........)))).)))))......... ( -30.70, z-score = -2.23, R) >droYak2.chr3R 28271245 92 - 28832112 GGCUGGAUGGU-------CAGCGGAUUUGGGUCACUUGGCUUGGGUCAGGCAACGGUUAUGAAACAAUUGCAGACAAUUAGCCAGCAGCCGGGGUAAAC (((((..((((-------.....((((..((((....))))..))))..((((..(((....)))..)))).........)))).)))))......... ( -30.70, z-score = -2.16, R) >droEre2.scaffold_4820 9827544 82 + 10470090 GGCUGGAUGGU-------CAGUGGAUUUGGGUCACUUGGCUUGGGUCAGGCAACGGUUAUGAAACAAUUGCAGACAAUUAGCCAGCAGC---------- .(((((...((-------(....((((..((((....))))..))))..((((..(((....)))..)))).)))......)))))...---------- ( -29.30, z-score = -2.96, R) >droAna3.scaffold_13340 16468177 99 - 23697760 GGCUGGAUGGUUAGCCGCCAGCAGAUUUGGGUCACUUGGCUUGGGUCAGGCAACGGUUAUGAAACAAUUGCAGACAAUUAGCCGGGAGUGGAGCUAAAC .(((((.(((....))))))))...(((((.((.(((((((...(((..((((..(((....)))..)))).)))....)))))))....)).))))). ( -36.80, z-score = -2.91, R) >dp4.chr2 26231741 84 + 30794189 GGCUGGAUGGC-------CAGAAGAUUUGGGUCACUUGGCUGGGGUCAGGCAACGGUUAUGAAACAAUUGCAGACAAUUAGCCAACGACCA-------- ((((....)))-------)..........((((..(((((((..(((..((((..(((....)))..)))).)))...))))))).)))).-------- ( -29.70, z-score = -2.66, R) >droPer1.super_19 274519 84 - 1869541 GGCUGGAUGGC-------CAGAAGAUUUGGGUCACUUGGCUGGGGUCAGGCAACGGUUAUGAAACAAUUGCAGACAAUUAGCCAACGACCA-------- ((((....)))-------)..........((((..(((((((..(((..((((..(((....)))..)))).)))...))))))).)))).-------- ( -29.70, z-score = -2.66, R) >consensus GGCUGGAUGGU_______CAGCGGAUUUGGGUCACUUGGCUUGGGUCAGGCAACGGUUAUGAAACAAUUGCAGACAAUUAGCCAGCAGCCGAGGUAAAC (((((..((((............((((..((((....))))..))))..((((..(((....)))..)))).........)))).)))))......... (-26.25 = -26.29 + 0.03)

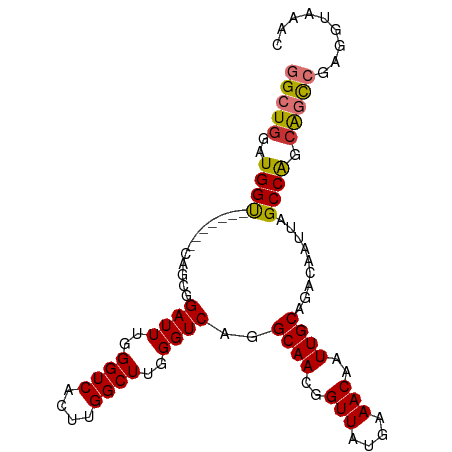
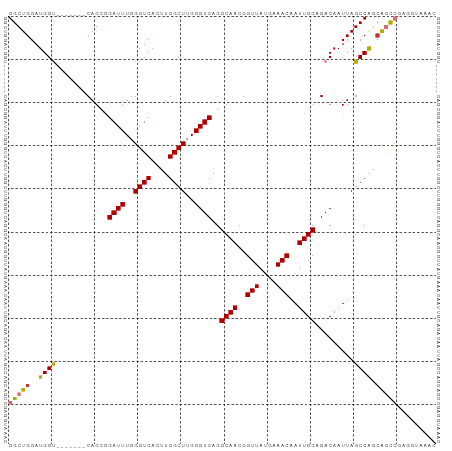
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:59:53 2011