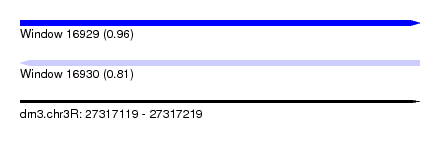
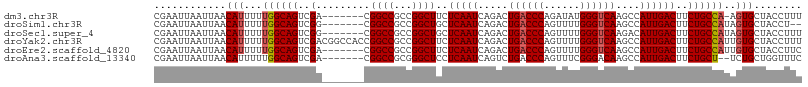
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 27,317,119 – 27,317,219 |
| Length | 100 |
| Max. P | 0.959378 |

| Location | 27,317,119 – 27,317,219 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 90.19 |
| Shannon entropy | 0.17968 |
| G+C content | 0.49340 |
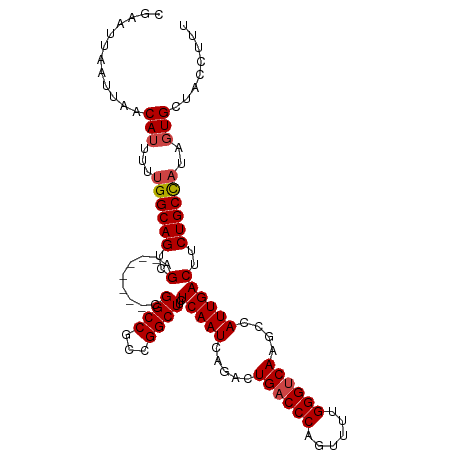
| Mean single sequence MFE | -34.17 |
| Consensus MFE | -27.92 |
| Energy contribution | -28.62 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.67 |
| SVM RNA-class probability | 0.959378 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
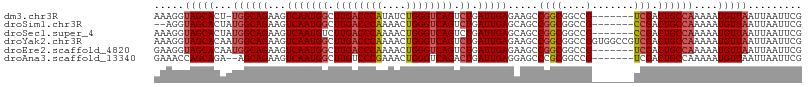
>dm3.chr3R 27317119 100 + 27905053 AAAGGUAGCACU-UGGCAGAAGUCAAUGGCUUGACCCAUAUCUGGGUCAGUCUGAUUGAGAAGCCGGCGGCCG-------UCGACUGCCAAAAAUGUUAAUUAAUUCG .....(((((.(-((((((.(((((..((((.((((((....))))))))))))))).......((((....)-------))).)))))))...)))))......... ( -34.10, z-score = -2.60, R) >droSim1.chr3R 26948798 99 + 27517382 --AGGUAGCACUAUGGCAGAAGUCAAUGGCUUGACCCAAAACUGGGUCAGUCUGAUUGAGCAGCCGGCGGCCG-------CCGACUGCCAAAAAUGUUAAUUAAUUCG --...(((((...((((((.(((((..((((.((((((....))))))))))))))).......((((....)-------))).))))))....)))))......... ( -36.10, z-score = -2.76, R) >droSec1.super_4 6116538 101 + 6179234 AAAGGUAGCACUAUGGCAGAAGUCAAUGUCUUGACCCAAAACUGGGUCAGUCUGAUUGAGCAGCCGGCGGCCG-------CCGACUGCCAAAAAUGUUAAUUAAUUCG .....(((((...((((((...(((((...((((((((....))))))))....))))).....((((....)-------))).))))))....)))))......... ( -33.60, z-score = -2.55, R) >droYak2.chr3R 28250173 108 + 28832112 AAAGGUAGCACAAUGGCAGAAGUCAAUGGCUUGACCCAAAACUGGGUCAGUCUGAUUGAGAAGCCGGCGGCCGGUGGCCGUCGACUGCCAAAAAUGUUAAUUAAUUCG .....(((((...((((((.(((((..((((.((((((....))))))))))))))).......((((((((...)))))))).))))))....)))))......... ( -43.70, z-score = -4.23, R) >droEre2.scaffold_4820 9802707 101 - 10470090 GAAGGUAGCACAAUGGCAGAAGUCAAUGGCUUGACCCAAAACUGGGUCAGUCUGAUUGAGAAGCCGGCGGCCG-------UCGACUGCCAAAAAUGUUAAUUAAUUCG (((..(((((...((((((.(((((..((((.((((((....))))))))))))))).......((((....)-------))).))))))....))))).....))). ( -35.00, z-score = -3.05, R) >droAna3.scaffold_13340 16444513 99 + 23697760 GAAACCAGCAGA--AGCAGAAGUCAAUGGCUUGUCCCGAAACUGGGUCAGACUGAUUGAGGAGCCCGCGGCCG-------UCGACUGCCAAAAAUGUUAAUUAAUUCG .......((((.--..(((((((.....))))(.((((....)))).)...))).((((.(.(((...)))).-------)))))))).................... ( -22.50, z-score = 0.50, R) >consensus AAAGGUAGCACAAUGGCAGAAGUCAAUGGCUUGACCCAAAACUGGGUCAGUCUGAUUGAGAAGCCGGCGGCCG_______UCGACUGCCAAAAAUGUUAAUUAAUUCG .....(((((...((((((...(((((((.((((((((....)))))))).)).)))))...(((...))).............))))))....)))))......... (-27.92 = -28.62 + 0.69)
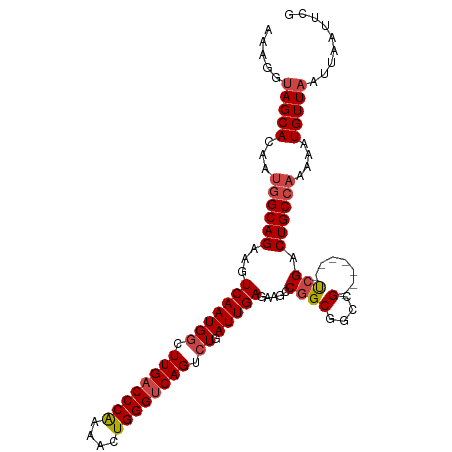
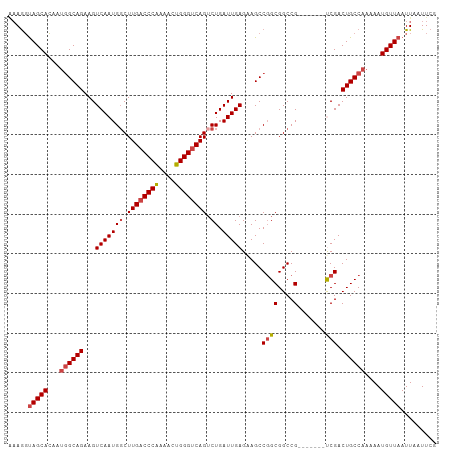
| Location | 27,317,119 – 27,317,219 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 90.19 |
| Shannon entropy | 0.17968 |
| G+C content | 0.49340 |
| Mean single sequence MFE | -30.62 |
| Consensus MFE | -24.47 |
| Energy contribution | -24.83 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.810488 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 27317119 100 - 27905053 CGAAUUAAUUAACAUUUUUGGCAGUCGA-------CGGCCGCCGGCUUCUCAAUCAGACUGACCCAGAUAUGGGUCAAGCCAUUGACUUCUGCCA-AGUGCUACCUUU ................((((((((((((-------.(((........(((.....))).(((((((....))))))).))).)))))...)))))-)).......... ( -28.80, z-score = -2.06, R) >droSim1.chr3R 26948798 99 - 27517382 CGAAUUAAUUAACAUUUUUGGCAGUCGG-------CGGCCGCCGGCUGCUCAAUCAGACUGACCCAGUUUUGGGUCAAGCCAUUGACUUCUGCCAUAGUGCUACCU-- ............((((..((((((..((-------(((((...))))))(((((..(..((((((......))))))..).))))))..)))))).))))......-- ( -33.90, z-score = -2.53, R) >droSec1.super_4 6116538 101 - 6179234 CGAAUUAAUUAACAUUUUUGGCAGUCGG-------CGGCCGCCGGCUGCUCAAUCAGACUGACCCAGUUUUGGGUCAAGACAUUGACUUCUGCCAUAGUGCUACCUUU ............((((..((((((..((-------(((((...))))))(((((.....((((((......))))))....))))))..)))))).))))........ ( -33.20, z-score = -2.66, R) >droYak2.chr3R 28250173 108 - 28832112 CGAAUUAAUUAACAUUUUUGGCAGUCGACGGCCACCGGCCGCCGGCUUCUCAAUCAGACUGACCCAGUUUUGGGUCAAGCCAUUGACUUCUGCCAUUGUGCUACCUUU ............(((...(((((((((.(((((...))))).))))...(((((..(..((((((......))))))..).)))))....)))))..)))........ ( -35.60, z-score = -3.10, R) >droEre2.scaffold_4820 9802707 101 + 10470090 CGAAUUAAUUAACAUUUUUGGCAGUCGA-------CGGCCGCCGGCUUCUCAAUCAGACUGACCCAGUUUUGGGUCAAGCCAUUGACUUCUGCCAUUGUGCUACCUUC ............(((...((((((((((-------.(((........(((.....))).((((((......)))))).))).)))))...)))))..)))........ ( -27.90, z-score = -1.74, R) >droAna3.scaffold_13340 16444513 99 - 23697760 CGAAUUAAUUAACAUUUUUGGCAGUCGA-------CGGCCGCGGGCUCCUCAAUCAGUCUGACCCAGUUUCGGGACAAGCCAUUGACUUCUGCU--UCUGCUGGUUUC .((((((............((.((((((-------.(((..((((((........)))))).(((......)))....))).)))))).))((.--...)))))))). ( -24.30, z-score = 0.16, R) >consensus CGAAUUAAUUAACAUUUUUGGCAGUCGA_______CGGCCGCCGGCUUCUCAAUCAGACUGACCCAGUUUUGGGUCAAGCCAUUGACUUCUGCCAUAGUGCUACCUUU ............(((...((((((..(.........((((...))))..(((((.....((((((......))))))....))))))..))))))..)))........ (-24.47 = -24.83 + 0.36)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:59:48 2011