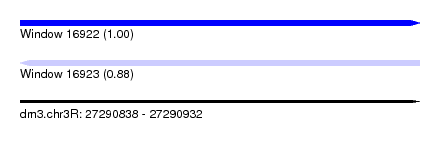
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 27,290,838 – 27,290,932 |
| Length | 94 |
| Max. P | 0.997043 |

| Location | 27,290,838 – 27,290,932 |
|---|---|
| Length | 94 |
| Sequences | 8 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 65.76 |
| Shannon entropy | 0.62451 |
| G+C content | 0.52285 |
| Mean single sequence MFE | -27.86 |
| Consensus MFE | -17.11 |
| Energy contribution | -16.68 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.03 |
| SVM RNA-class probability | 0.997043 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
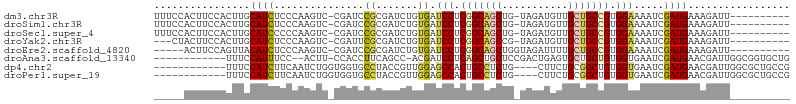
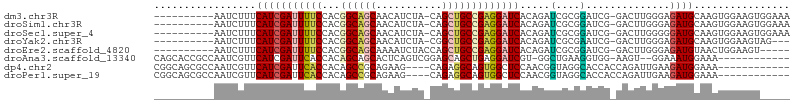
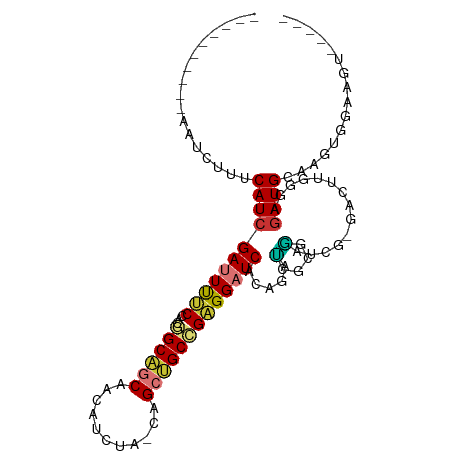
>dm3.chr3R 27290838 94 + 27905053 UUUCCACUUCCACUUGCAUCUCCCAAGUC-CGAUCCGCGAUCUGUGAUCCUCGGCAGCUG-UAGAUGUUGCUGCCGUGGAAAAUCGAUGAAAGAUU---------- ............(((.((((((.((..((-((...)).))..)).))(((.(((((((..-........))))))).))).....)))).)))...---------- ( -27.40, z-score = -2.05, R) >droSim1.chr3R 26922292 94 + 27517382 UUUCCACUUCCACUUGCAUCUCCCAAGUC-CGAUCCGCGAUCUGUGAUCCUCGGCAGCUG-UAGAUGUUGCUGCCGUGGAAAAUCGAUGAAAGAUU---------- ............(((.((((((.((..((-((...)).))..)).))(((.(((((((..-........))))))).))).....)))).)))...---------- ( -27.40, z-score = -2.05, R) >droSec1.super_4 6090527 94 + 6179234 UUUCCACUUCCACUUGCAUCCCCCAAGUC-CGAUCCGCGAUCUGUGAUCCUCGGCAGCUG-UAGAUGUUGCUGCCGUGGAAAAUCGAUGAAAGAUU---------- ............(((.((((.........-.((((...)))).....(((.(((((((..-........))))))).))).....)))).)))...---------- ( -26.10, z-score = -1.90, R) >droYak2.chr3R 28223400 91 + 28832112 ---CUACUUCCACUUGCAUCUCCCAAGUC-CGAUUCGCGAUCUGUGAUCCUCGGCAGCCG-UAGAUGUUGCUGCCGUGGAAAAUCGAUGAAAGAUU---------- ---.........(((.((((((.((..((-((...)).))..)).))(((.(((((((..-........))))))).))).....)))).)))...---------- ( -26.20, z-score = -1.80, R) >droEre2.scaffold_4820 9776488 90 - 10470090 -----ACUUCCAGUUACAUCUCCCAAGUC-CGAUCCGCGAUCUGUGAUCCUCGGCAGCUGGUAGAUUUUGCUGCCGUGGAAAAUCGAUGAAAGAUU---------- -----...........((((((.((..((-((...)).))..)).))(((.(((((((...........))))))).))).....)))).......---------- ( -25.40, z-score = -1.41, R) >droAna3.scaffold_13340 16416172 90 + 23697760 ------------UUUCCAUUUCC--ACUU-CCACCUUCAGCC-ACGAUCCUCAGCUGCUCCGACUGAGUGCUGCUGUGGUGAAUCGAUGAACGAUUGGCGGUGCUG ------------..........(--(((.-((.......(((-(((.....((((.((((.....)))))))).)))))).(((((.....))))))).))))... ( -24.00, z-score = -0.48, R) >dp4.chr2 4516765 90 + 30794189 ------------UUUCCAUCUUCAAUCUGGUGGUGCCUACCGUUGGAGCCACUGCCUCUG----CUUCUGCGGCUGUGGUGAAUCGAUGAACGAUUGGCGCUGCCG ------------................((..(((((...((((((.(((((.(((...(----(....))))).)))))...)))))).......)))))..)). ( -33.20, z-score = -2.64, R) >droPer1.super_19 215552 90 + 1869541 ------------UUUCCAUCUUCAAUCUGGUGGUGCCUACCGUUGGAGCCACUGCCUCUG----CUUCUGCGGCUGUGGUGAAUCGAUGAACGAUUGGCGCUGCCG ------------................((..(((((...((((((.(((((.(((...(----(....))))).)))))...)))))).......)))))..)). ( -33.20, z-score = -2.64, R) >consensus _____ACUUCCACUUGCAUCUCCCAAGUC_CGAUCCGCGAUCUGUGAUCCUCGGCAGCUG_UAGAUGUUGCUGCCGUGGAAAAUCGAUGAAAGAUU__________ ................((((...............((.......)).(((.(((((((...........))))))).))).....))))................. (-17.11 = -16.68 + -0.44)
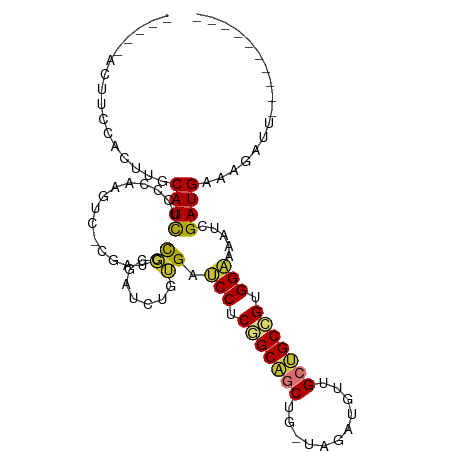
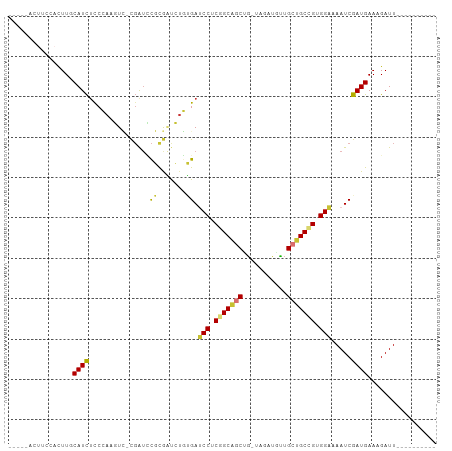
| Location | 27,290,838 – 27,290,932 |
|---|---|
| Length | 94 |
| Sequences | 8 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 65.76 |
| Shannon entropy | 0.62451 |
| G+C content | 0.52285 |
| Mean single sequence MFE | -27.39 |
| Consensus MFE | -13.90 |
| Energy contribution | -13.70 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.12 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.876626 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 27290838 94 - 27905053 ----------AAUCUUUCAUCGAUUUUCCACGGCAGCAACAUCUA-CAGCUGCCGAGGAUCACAGAUCGCGGAUCG-GACUUGGGAGAUGCAAGUGGAAGUGGAAA ----------...(((((((.((((((((.(((((((........-..))))))).)))....)))))(((..((.-.......))..)))..)))))))...... ( -30.00, z-score = -1.75, R) >droSim1.chr3R 26922292 94 - 27517382 ----------AAUCUUUCAUCGAUUUUCCACGGCAGCAACAUCUA-CAGCUGCCGAGGAUCACAGAUCGCGGAUCG-GACUUGGGAGAUGCAAGUGGAAGUGGAAA ----------...(((((((.((((((((.(((((((........-..))))))).)))....)))))(((..((.-.......))..)))..)))))))...... ( -30.00, z-score = -1.75, R) >droSec1.super_4 6090527 94 - 6179234 ----------AAUCUUUCAUCGAUUUUCCACGGCAGCAACAUCUA-CAGCUGCCGAGGAUCACAGAUCGCGGAUCG-GACUUGGGGGAUGCAAGUGGAAGUGGAAA ----------...(((((((.((((((((.(((((((........-..))))))).)))....)))))(((..((.-.......))..)))..)))))))...... ( -29.10, z-score = -1.37, R) >droYak2.chr3R 28223400 91 - 28832112 ----------AAUCUUUCAUCGAUUUUCCACGGCAGCAACAUCUA-CGGCUGCCGAGGAUCACAGAUCGCGAAUCG-GACUUGGGAGAUGCAAGUGGAAGUAG--- ----------...(((((((.((((((((.(((((((........-..))))))).)))....)))))(((..((.-.......))..)))..)))))))...--- ( -28.70, z-score = -1.95, R) >droEre2.scaffold_4820 9776488 90 + 10470090 ----------AAUCUUUCAUCGAUUUUCCACGGCAGCAAAAUCUACCAGCUGCCGAGGAUCACAGAUCGCGGAUCG-GACUUGGGAGAUGUAACUGGAAGU----- ----------.((((..((((((...(((.(((((((...........)))))))..((((...))))..))))))-)...))..))))............----- ( -27.80, z-score = -1.73, R) >droAna3.scaffold_13340 16416172 90 - 23697760 CAGCACCGCCAAUCGUUCAUCGAUUCACCACAGCAGCACUCAGUCGGAGCAGCUGAGGAUCGU-GGCUGAAGGUGG-AAGU--GGAAAUGGAAA------------ ((...((((...(((.....)))((((((.((((.((.((((((.......))))))....))-.))))..)))))-).))--))...))....------------ ( -27.50, z-score = -0.53, R) >dp4.chr2 4516765 90 - 30794189 CGGCAGCGCCAAUCGUUCAUCGAUUCACCACAGCCGCAGAAG----CAGAGGCAGUGGCUCCAACGGUAGGCACCACCAGAUUGAAGAUGGAAA------------ .(((...))).....((((((...(((((((.(((((....)----)...))).)))).......(((.(....))))....))).))))))..------------ ( -23.00, z-score = 0.07, R) >droPer1.super_19 215552 90 - 1869541 CGGCAGCGCCAAUCGUUCAUCGAUUCACCACAGCCGCAGAAG----CAGAGGCAGUGGCUCCAACGGUAGGCACCACCAGAUUGAAGAUGGAAA------------ .(((...))).....((((((...(((((((.(((((....)----)...))).)))).......(((.(....))))....))).))))))..------------ ( -23.00, z-score = 0.07, R) >consensus __________AAUCUUUCAUCGAUUUUCCACGGCAGCAACAUCUA_CAGCUGCCGAGGAUCACAGAUCGCGGAUCG_GACUUGGGAGAUGCAAGUGGAAGU_____ .................(((((((((((...((((((...........)))))))))))))......(.(((........))).).))))................ (-13.90 = -13.70 + -0.20)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:59:43 2011