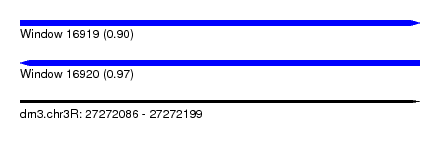
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 27,272,086 – 27,272,199 |
| Length | 113 |
| Max. P | 0.968550 |

| Location | 27,272,086 – 27,272,199 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 86.67 |
| Shannon entropy | 0.22799 |
| G+C content | 0.34298 |
| Mean single sequence MFE | -21.26 |
| Consensus MFE | -16.14 |
| Energy contribution | -15.54 |
| Covariance contribution | -0.60 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.900183 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
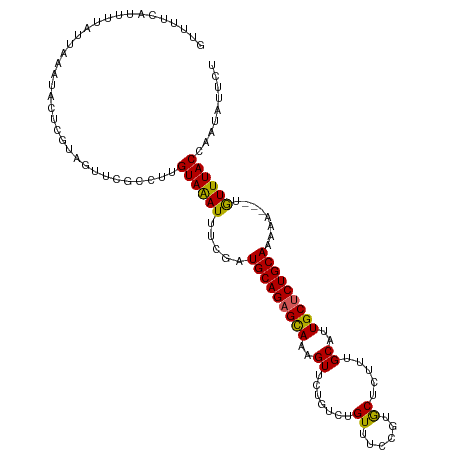
>dm3.chr3R 27272086 113 + 27905053 GUUUUCAUUUUAUUAAAUACUCGUAGUUCGCCUUGUAAAUUUCGAUGCAGAGCAAAGUUUUGUCUGUUUCCGUACGCUUUGCAUUGCUCUGCAAAAA---UGUUUACCAAUAUUCU ((...((((((.........(((.((((..(...)..)))).)))((((((((((.((...((.((......)).))...)).))))))))))))))---))...))......... ( -24.20, z-score = -2.66, R) >droSim1.chr3R 26902417 114 + 27517382 GUUUCCAUUUUAUUAAAUACUCGUAGUUCGCCUUGUAAAUUCCGAUGCAGAGCAAAGUU--GUCUGUUUCCGUGCUCUUUGCAUUGCUCUGCAAAAAAAAUGUUUACCAAUAUUCU ((...((((((.........(((.((((..(...)..)))).)))((((((((((.((.--....((......)).....)).))))))))))...))))))...))......... ( -24.10, z-score = -3.21, R) >droSec1.super_4 6072075 116 + 6179234 GUUUCCAUUUUAUUAAAUACUCGUAGUUCGCCUUGUAGAUUCCGAUGCAGAGCAUAGUUUUGUCUGUUUCCGUGCACUUUGCAUUGCUCUGCAAAAAAAAUGUUUACCAAUAUUCU ((...((((((.........(((.((((..(...)..)))).)))((((((((((((......))))....((((.....)))).))))))))...))))))...))......... ( -24.60, z-score = -2.52, R) >droYak2.chr3R 28202104 107 + 28832112 GUUUUCAUUUUAUUAAAUA---GUUCGUC---UUGUAAAUUUCAAUGCAGAGUAAAGUUCUGUCUGUUUCCAUGCUCUUUGCACUGCGCUGCAAAAA---UAUUUACCAAUAUUCU ...................---.......---..((((((..((.(((((.((((((....((.((....)).)).)))))).))))).))......---.))))))......... ( -15.10, z-score = -0.53, R) >droEre2.scaffold_4820 9757847 109 - 10470090 -GUUUCAUUUUAUUAAAUA---GUUCGUCGUCUUGUAAAUUUCGAUGCAGAGUAAAGUUCUGUCUGUUUCCUUGCUCUUUGCACUGCUCUGCAAAAA---UAUUUACCAAUAUUCU -..................---............((((((.....(((((((((.((......)).......(((.....))).)))))))))....---.))))))......... ( -18.30, z-score = -2.02, R) >consensus GUUUUCAUUUUAUUAAAUACUCGUAGUUCGCCUUGUAAAUUUCGAUGCAGAGCAAAGUUCUGUCUGUUUCCGUGCUCUUUGCAUUGCUCUGCAAAAA___UGUUUACCAAUAUUCU ..................................((((((.....(((((((((..((.......((......)).....))..)))))))))........))))))......... (-16.14 = -15.54 + -0.60)

| Location | 27,272,086 – 27,272,199 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 86.67 |
| Shannon entropy | 0.22799 |
| G+C content | 0.34298 |
| Mean single sequence MFE | -23.76 |
| Consensus MFE | -18.26 |
| Energy contribution | -19.58 |
| Covariance contribution | 1.32 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.80 |
| SVM RNA-class probability | 0.968550 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
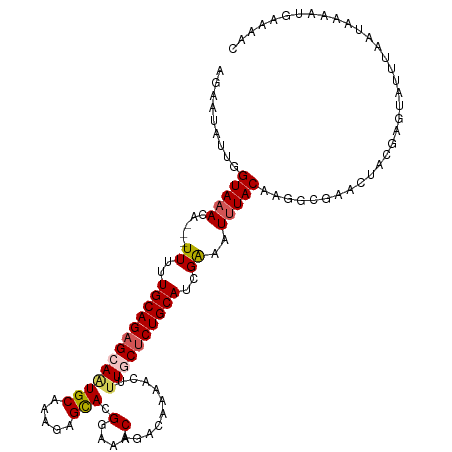
>dm3.chr3R 27272086 113 - 27905053 AGAAUAUUGGUAAACA---UUUUUGCAGAGCAAUGCAAAGCGUACGGAAACAGACAAAACUUUGCUCUGCAUCGAAAUUUACAAGGCGAACUACGAGUAUUUAAUAAAAUGAAAAC .(((((((.(((....---....((((((((((.((...))((..(....)..))......))))))))))(((............)))..))).))))))).............. ( -26.10, z-score = -3.04, R) >droSim1.chr3R 26902417 114 - 27517382 AGAAUAUUGGUAAACAUUUUUUUUGCAGAGCAAUGCAAAGAGCACGGAAACAGAC--AACUUUGCUCUGCAUCGGAAUUUACAAGGCGAACUACGAGUAUUUAAUAAAAUGGAAAC .(((((((.(((...........(((((((((((((.....))).(....)....--....))))))))))(((............)))..))).))))))).............. ( -27.60, z-score = -3.54, R) >droSec1.super_4 6072075 116 - 6179234 AGAAUAUUGGUAAACAUUUUUUUUGCAGAGCAAUGCAAAGUGCACGGAAACAGACAAAACUAUGCUCUGCAUCGGAAUCUACAAGGCGAACUACGAGUAUUUAAUAAAAUGGAAAC .(((((((.(((...........(((((((((.(((.....))).(....)...........)))))))))(((............)))..))).))))))).............. ( -27.30, z-score = -2.98, R) >droYak2.chr3R 28202104 107 - 28832112 AGAAUAUUGGUAAAUA---UUUUUGCAGCGCAGUGCAAAGAGCAUGGAAACAGACAGAACUUUACUCUGCAUUGAAAUUUACAA---GACGAAC---UAUUUAAUAAAAUGAAAAC .........((((((.---((..(((((.(..((((.....))))(....).............).)))))..)).))))))..---.......---................... ( -17.70, z-score = -0.99, R) >droEre2.scaffold_4820 9757847 109 + 10470090 AGAAUAUUGGUAAAUA---UUUUUGCAGAGCAGUGCAAAGAGCAAGGAAACAGACAGAACUUUACUCUGCAUCGAAAUUUACAAGACGACGAAC---UAUUUAAUAAAAUGAAAC- .........((((((.---((..(((((((...(((.....))).(....).............)))))))..)).))))))............---..................- ( -20.10, z-score = -2.22, R) >consensus AGAAUAUUGGUAAACA___UUUUUGCAGAGCAAUGCAAAGAGCACGGAAACAGACAAAACUUUGCUCUGCAUCGAAAUUUACAAGGCGAACUACGAGUAUUUAAUAAAAUGAAAAC .(((((((.((............(((((((((((((.....))).(....)..........))))))))))(((............)))...)).))))))).............. (-18.26 = -19.58 + 1.32)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:59:40 2011