| Sequence ID | dm3.chr3R |
|---|---|
| Location | 27,245,379 – 27,245,479 |
| Length | 100 |
| Max. P | 0.783446 |

| Location | 27,245,379 – 27,245,479 |
|---|---|
| Length | 100 |
| Sequences | 10 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 77.11 |
| Shannon entropy | 0.43146 |
| G+C content | 0.37768 |
| Mean single sequence MFE | -21.74 |
| Consensus MFE | -10.20 |
| Energy contribution | -10.20 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.783446 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
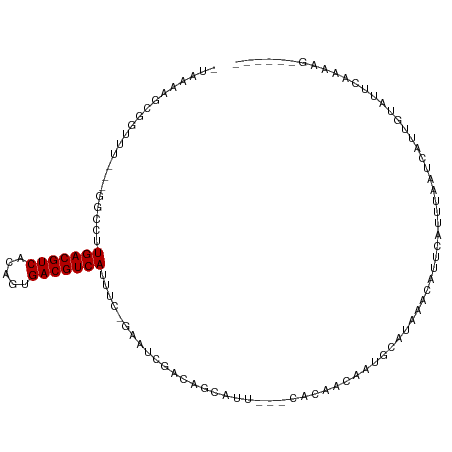
>dm3.chr3R 27245379 100 - 27905053 -UAAAAGCGGUUU---GUGCUUGACGUCACAGUGACGUCAUUUC-GAAUCGACAGCAUUCACCACAUCGCUGCAUAAACAUUCAUUUAAUCAUUCCAUUCCGAAG------ -.....(((((.(---(((..(((((((.....)))))))....-(((((....).))))..))))..)))))................................------ ( -22.40, z-score = -1.91, R) >droPer1.super_19 145636 90 - 1869541 -UAAAAGCGGUUUCGUUGCUUUGACGUCACUGUGACGUCAUUUG-GCAU--ACAA-----------ACAAUGCAUAAACAUUCGUUUAAUCAUUGUAUUUAAAAG------ -.............((((((.(((((((.....)))))))...)-))).--))..-----------((((((..(((((....)))))..)))))).........------ ( -23.70, z-score = -3.04, R) >dp4.chr2 4443181 90 - 30794189 -UAAAAGCGGUUUCGUUGCUUUGACGUCACUGUGACGUCAUUUG-GCAU--ACAA-----------ACAAUGCAUAAACAUUCGUUUAAUCAUUGUAUUUAAAAG------ -.............((((((.(((((((.....)))))))...)-))).--))..-----------((((((..(((((....)))))..)))))).........------ ( -23.70, z-score = -3.04, R) >droEre2.scaffold_4820 9728496 98 + 10470090 AUAAAAGCGGUUU---GUGCUUGACGUCACAGUGACGUCAUUUC-GAAUCGACAGCAUU---CACAUCGAUGCAUAAACAUUCACUUAAUCAUUAUAUUCCAAAG------ ......(((((((---(....(((((((.....)))))))...)-)))))).).(((((---......)))))................................------ ( -22.80, z-score = -2.48, R) >droYak2.chr3R 28172013 98 - 28832112 AUAAAAGCGGUUU---GUGCUUGACGUCACAGUGACGUCAUUUC-GCACCGACAGCAUU---CACAUCGAUGCAUAAACAUUCAUUUAAUCAUUAUAUUCCGAAG------ .......(((((.---((((.(((((((.....)))))))....-)))).))).(((((---......)))))...........................))...------ ( -23.60, z-score = -2.35, R) >droSec1.super_4 6045370 100 - 6179234 -UAAAAGCGGUUU---GUGCUUGACGUCACAGUGACGUCAUUUC-GAAUCGACAGCAUUCACCACAUCGCUGCAUAAACAUUCAUUUAAUCAUUCCAUACCGAAG------ -.....(((((.(---(((..(((((((.....)))))))....-(((((....).))))..))))..)))))................................------ ( -22.40, z-score = -1.95, R) >droSim1.chr3R 26875591 100 - 27517382 -UAAAAGCGGUUU---GUGCUUGACGUCACAGUGACGUCAUUUC-GAAUCGACAGCAUUCACCACAUCGCUGCAUAAACAUUCAUUUAAUCAUUCCAUUCCGAAG------ -.....(((((.(---(((..(((((((.....)))))))....-(((((....).))))..))))..)))))................................------ ( -22.40, z-score = -1.91, R) >droWil1.scaffold_181108 3282481 104 - 4707319 -AAAAAGCGGUCU----GCUUUGACGUCAUAGUGACGUCAUUUG-GAAUUGACAAGCUA-GAGAACACAAUGCAUAAACAUUCAUUUAAUCAUUGUAUUUAAAAAAAAAAA -....(((.(((.----.((.(((((((.....)))))))...)-)....)))..))).-..(((.((((((..((((......))))..)))))).)))........... ( -21.70, z-score = -1.92, R) >droMoj3.scaffold_6540 4956001 94 + 34148556 -UAAAAGCGGAUC----GCUUUGACGUCACUGUGACGUCAUUUGCACAACGACAGC------AAUAACAAGGCAUAAACAUUAACUUAAUCAUUGAAUUUAAGAG------ -............----(((((((((((.....))))))..((((.........))------))....)))))...........(((((.........)))))..------ ( -17.70, z-score = -1.31, R) >droVir3.scaffold_13047 12644177 94 - 19223366 -UAAAAGCGGAUC----GCUUUGACGUCACUGUGACGUCAUUUGGGCAUCGACAGC------AAUAACAAGGCAUAAACAUUAACUUAAUCAUUGUAUUUAAAAG------ -((((.((((((.----(((((((((((.....)))))))...)))))))....((------.........))....................))).))))....------ ( -17.00, z-score = -0.80, R) >consensus _UAAAAGCGGUUU___GGCCUUGACGUCACAGUGACGUCAUUUC_GAAUCGACAGCAUU___CACAACAAUGCAUAAACAUUCAUUUAAUCAUUGUAUUCAAAAG______ .....................(((((((.....)))))))....................................................................... (-10.20 = -10.20 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:59:37 2011