| Sequence ID | dm3.chr2L |
|---|---|
| Location | 9,338,814 – 9,338,876 |
| Length | 62 |
| Max. P | 0.500000 |

| Location | 9,338,814 – 9,338,876 |
|---|---|
| Length | 62 |
| Sequences | 11 |
| Columns | 62 |
| Reading direction | reverse |
| Mean pairwise identity | 75.54 |
| Shannon entropy | 0.57097 |
| G+C content | 0.42556 |
| Mean single sequence MFE | -14.39 |
| Consensus MFE | -7.35 |
| Energy contribution | -7.59 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.71 |
| Mean z-score | -0.87 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
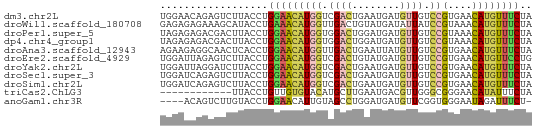
>dm3.chr2L 9338814 62 - 23011544 UGGAACAGAGUCUUACCUGGAACAUGGUCGACUGAAUGAUGUUGUCCGUGAACAUGUUUCUA .((((((...(((.....))).(((((.((((........)))).)))))....)))))).. ( -17.20, z-score = -1.47, R) >droWil1.scaffold_180708 426599 62 - 12563649 GAGAGAGAAAGCAUACCUGAAACAUGGUUGACUGUAUGAUAUUAUCCGUAAACAUGUUUCUA ..................((((((((.(((...(((......)))...))).)))))))).. ( -9.80, z-score = -0.58, R) >droPer1.super_5 4235125 62 - 6813705 UAGAGAGACGACUUACCUGGAACAUGGUGGACUGGAUGAUGUUGUCCGUAAACAUGUUUCUA ...(((((((..(((((........)))))...((((......)))).......))))))). ( -14.50, z-score = -0.80, R) >dp4.chr4_group1 4305088 62 + 5278887 UAGAGAGACGACUUACCUGGAACAUGGUGGACUGGAUGAUGUUGUCCGUAAACAUGUUUCUA ...(((((((..(((((........)))))...((((......)))).......))))))). ( -14.50, z-score = -0.80, R) >droAna3.scaffold_12943 678959 62 - 5039921 AGAAGAGGCAACUCACCUGGAACAUGGUUGACUGAAUUAUGUUGUCCGUGAACAUGUUUCUA ...(((((((..((((..(.(((((((((.....))))))))).)..))))...))))))). ( -17.40, z-score = -1.80, R) >droEre2.scaffold_4929 9949048 62 - 26641161 UGGAUUAGAGUCUUACCUGGAACAUGGUCGACUGUAUGAUGUUGUCCGUGAACAUGUUCCUG .((((....)))).....(((((((((.((((........)))).))(....)))))))).. ( -16.60, z-score = -1.39, R) >droYak2.chr2L 12008919 62 - 22324452 UGGAUUAGGAUCUUACCUGGAACAUGGUCGACUGAAUGAUGUUGUCCGUGAACAUGUUUCUA (((((((((......)))))..(((((.((((........)))).)))))........)))) ( -15.70, z-score = -1.17, R) >droSec1.super_3 4796676 62 - 7220098 UGGAUCAGAGUCUUACCUGGAACAUGGUCGACUGAAUGAUGUUGUCCGUGAACAUGUUUCUA ((((((((........))))..(((((.((((........)))).)))))........)))) ( -14.70, z-score = -0.34, R) >droSim1.chr2L 9116099 62 - 22036055 UGGAUCAGAGUCUUACCUGGAACAUGGUCGACUGAAUGAUGUUGUCCGUGAACAUGUUUCUA ((((((((........))))..(((((.((((........)))).)))))........)))) ( -14.70, z-score = -0.34, R) >triCas2.ChLG3 18084270 50 - 32080666 ------------UUACCUGUUGUGUACAUGCUUGAAUGACGUUGGGCGGGAACAUAUUUCUA ------------........(((((.(.((((..(......)..)))).).)))))...... ( -9.50, z-score = -0.36, R) >anoGam1.chr3R 34092601 57 - 53272125 ----ACAGUCUUGUACCUGGAACAUUGUAGCCUGGAUGAUGUUCGGUGGGAAUAGAUUUCU- ----..(((((....(((.((((((..(.......)..)))))).).))....)))))...- ( -13.70, z-score = -0.56, R) >consensus UGGAGAAGAGUCUUACCUGGAACAUGGUCGACUGAAUGAUGUUGUCCGUGAACAUGUUUCUA ..................(((((((((.((((........)))).))(....)))))))).. ( -7.35 = -7.59 + 0.24)
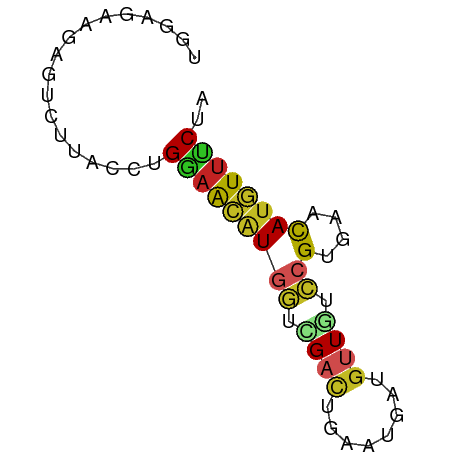
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:28:00 2011