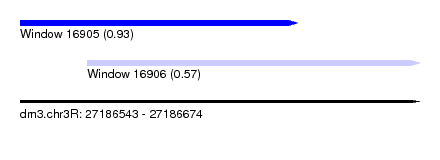
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 27,186,543 – 27,186,674 |
| Length | 131 |
| Max. P | 0.934040 |

| Location | 27,186,543 – 27,186,634 |
|---|---|
| Length | 91 |
| Sequences | 13 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 62.77 |
| Shannon entropy | 0.73360 |
| G+C content | 0.44261 |
| Mean single sequence MFE | -22.57 |
| Consensus MFE | -13.38 |
| Energy contribution | -13.32 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.36 |
| Mean z-score | -0.55 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.934040 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
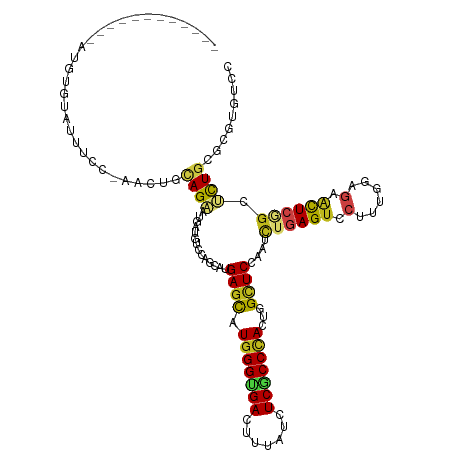
>dm3.chr3R 27186543 91 + 27905053 -------------------------CUUCACAAAUUUU-GGAAUAUGAAUGUGGAUUUCCGAACU-UCAGAAUGAUGUCCAGCAUGAGCAUGGGUGACUUUAUCUCGCCCACUGGCUC -------------------------...........((-(((...((((..(((....)))...)-)))........)))))...((((.(((((((.......)))))))...)))) ( -23.20, z-score = -0.79, R) >droSim1.chr3R 26821149 90 + 27517382 -------------------------CUUUACAAAUUUG-GGAG-AUGAAUGUGGAUUUUCCAACU-UCAGAAUGAUGUCCAGCAUGAGCAUGGGUGACUUUAUCUCGCCCACUGGCUC -------------------------..........(((-((..-.((((..(((.....)))..)-)))........)))))...((((.(((((((.......)))))))...)))) ( -23.40, z-score = -0.57, R) >droSec1.super_4 5991865 90 + 6179234 -------------------------CUUUACAAAUUUG-GGAA-AUUAAUGUAAAUUUCUCAACU-UCAGAAUGAUGUCCAGCAUGAGCAUGGGUGACUUUAUCUCGCCCACUGGCUC -------------------------..........(((-((((-(((......))))))))))..-........(((.....)))((((.(((((((.......)))))))...)))) ( -23.40, z-score = -2.07, R) >droYak2.chr3R 28116881 90 + 28832112 -------------------------GCUUACAACUUUG-GUAA-CUAUAUGUGGAUUACCCAACU-GCAGAAUGAUGAGCAGCAUGAGCAUGGGCGACUUCAUCUCGCCCACUGGCUC -------------------------(((((((.....(-((((-(((....))).)))))...(.-...)..)).))))).....((((.(((((((.......)))))))...)))) ( -25.50, z-score = -1.38, R) >droEre2.scaffold_4820 9671489 90 - 10470090 -------------------------CCUUGCAAAUGUG-GGAA-AUGUAUGUGGAUUAUCCAACU-UCAGAAUGAUGUCCAGCAUGAGCAUGGGUGACUUUAUCUCGCCCACUGGCUC -------------------------.((..(....)..-))..-....((((((((.(((.....-.......)))))))).)))((((.(((((((.......)))))))...)))) ( -24.50, z-score = -0.51, R) >droAna3.scaffold_13340 5065050 86 + 23697760 ------------------------------UUGGUUU-CAAAGUUAAAGGAUAUACGUGAUU-CGCCUAGAAUGAUGUCCAGCAUGAGCAUGGGUGACUUUAUUUCGCCCACGGGCUC ------------------------------.......-..........((((((.....(((-(.....)))).)))))).....((((.(((((((.......)))))))...)))) ( -22.70, z-score = -0.83, R) >dp4.chr2 4380227 88 + 30794189 --------------------------CUUCACAGGAUUCGAAGUAAGAGUAU-CACUUUCUU---UAUAGAAUGAUGUCUAGCAUGAGCAUGGGUGACUUCAUCUCGCCCACUGGUUC --------------------------.....(((........((.(((.(((-((..((((.---...)))))))))))).)).......(((((((.......)))))))))).... ( -21.00, z-score = -0.34, R) >droPer1.super_19 84055 88 + 1869541 --------------------------CUUCUGAGGAUUCGGAUUAAGAGUCU-UACUUUCUU---UAUAGAAUGAUGUCUAGCAUGAGCAUGGGUGACUUCAUCUCGCCCACUGGUUC --------------------------......(((((((.......))))))-).(.((((.---...)))).)(((.....)))((((.(((((((.......)))))))...)))) ( -20.40, z-score = 0.21, R) >droWil1.scaffold_181108 3223132 95 + 4707319 -----------------------AAAUAAAAGAUCUUGUGGAGACUAUAUAUGCAUUUUUCGUUUGCCAGAAUGAUGUCAAGCAUGAGUAUGGGUGAUUUUAUCUCACCCACUGGCUC -----------------------(((..(((((((.((((....).)))...).))))))..)))(((((..(.(((.....))).)...(((((((.......)))))))))))).. ( -24.00, z-score = -1.43, R) >droVir3.scaffold_13047 12571389 104 + 19223366 --------------GGAUCAAGAUCUUUCUUUGAGUAUUGUCCAACUCUCCCAUGUUUUAUCUUCUGCAGCAUGAUGUCCAGCAUGAGCAUGGGCGACUUUAUCUCACCUACGGGCUC --------------(((.(((...(((.....)))..))))))...((.(((((((((...((...(((......)))..))...))))))))).))..........((....))... ( -24.60, z-score = -0.27, R) >droGri2.scaffold_14830 5736832 117 - 6267026 AUCCAUUUACAGUUUAAUCAUGAAUCAUCGUUAUUUAUU-UCUACAUCUUUCAUCUCUUAUUAUUCCCAGCAUGAUGUCUAGCAUGAGCAUGGGCGAUUUUAUCUCACCCACUGGCUC .........((((....(((((...((((((........-...............................)))))).....)))))....(((.((.......)).))))))).... ( -16.33, z-score = 0.09, R) >droMoj3.scaffold_6540 4894294 91 - 34148556 --------------------------GAAUUCACAUCUC-UCCAUAUCUCGUUUCUUUGAUCGCUCGCAGCAUGAUGUCUAGCAUGAGCAUGGGUGACUUUAUCUCGCCCACCGGCUC --------------------------......(((((..-........(((......)))..((.....))..))))).......((((.(((((((.......)))))))...)))) ( -20.40, z-score = -0.22, R) >anoGam1.chr2R 55674837 95 - 62725911 -----------------------CUCUGGAUGAGUGCGCAAUGUACAAGGAUCUUCUCGUUUGCUUGCAGAAUGAUGUCCAGCAUGAGCAUGGGUGACUUCAUCUCGCCAACUGGAUC -----------------------.((..(....((((...((((....((((..((...((((....))))..)).)))).))))..)))).(((((.......)))))..)..)).. ( -24.00, z-score = 0.97, R) >consensus _________________________CUUUAUAAAUUUG_GGAGUAUGAAUGUGUAUUUUCUAACU_GCAGAAUGAUGUCCAGCAUGAGCAUGGGUGACUUUAUCUCGCCCACUGGCUC ..........................................................................(((.....)))((((.(((((((.......)))))))...)))) (-13.38 = -13.32 + -0.06)

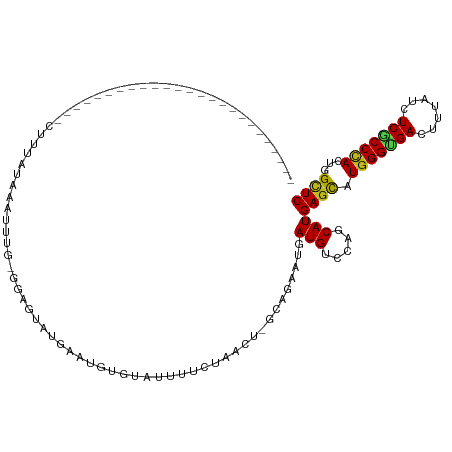
| Location | 27,186,565 – 27,186,674 |
|---|---|
| Length | 109 |
| Sequences | 13 |
| Columns | 121 |
| Reading direction | forward |
| Mean pairwise identity | 75.54 |
| Shannon entropy | 0.53635 |
| G+C content | 0.49957 |
| Mean single sequence MFE | -32.38 |
| Consensus MFE | -18.72 |
| Energy contribution | -18.88 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.55 |
| Mean z-score | -0.83 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.573082 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 27186565 109 + 27905053 ------------AUGUGGAUUUCCGAACUUCAGAAUGAUGUCCAGCAUGAGCAUGGGUGACUUUAUCUCGCCCACUGGCUCCAAUCUGAGUCCUUUGGAAAACUCAGCUCUGCGGGUGUCC ------------....(((((((((((.((((((.(((((.....)))((((.(((((((.......)))))))...)))))).))))))...))))))))((((.((...)))))).))) ( -34.50, z-score = -1.15, R) >droSim1.chr3R 26821170 109 + 27517382 ------------AUGUGGAUUUUCCAACUUCAGAAUGAUGUCCAGCAUGAGCAUGGGUGACUUUAUCUCGCCCACUGGCUCCAAUCUGAGUCCUUUGGAGAACUCGGCUCUGCGGGUGUCC ------------....(((((((((((.((((((.(((((.....)))((((.(((((((.......)))))))...)))))).))))))....)))))))(((((......))))))))) ( -36.30, z-score = -1.18, R) >droSec1.super_4 5991886 109 + 6179234 ------------AUGUAAAUUUCUCAACUUCAGAAUGAUGUCCAGCAUGAGCAUGGGUGACUUUAUCUCGCCCACUGGCUCCAAUCUGAGUCCUUUGGAGAACUCGGCUCUGCGGGUGUCC ------------........................(((..((.(((.((((.(((((((.......)))))))....((((((..........))))))......)))))))))..))). ( -33.80, z-score = -1.31, R) >droYak2.chr3R 28116902 109 + 28832112 ------------AUGUGGAUUACCCAACUGCAGAAUGAUGAGCAGCAUGAGCAUGGGCGACUUCAUCUCGCCCACUGGCUCCAAUCUGAGUCCUUUGGAGAACUCGGCUCUGCGCGUGUCC ------------...(((.....)))((((((((.....((((.((....)).(((((((.......)))))))...))))....((((((.((....)).)))))).)))))).)).... ( -35.10, z-score = -0.38, R) >droEre2.scaffold_4820 9671510 109 - 10470090 ------------AUGUGGAUUAUCCAACUUCAGAAUGAUGUCCAGCAUGAGCAUGGGUGACUUUAUCUCGCCCACUGGCUCCAAUCUGAGUCCUUUGGAGAACUCGGCUCUGCGAGUGUCC ------------....((((..(((((.((((((.(((((.....)))((((.(((((((.......)))))))...)))))).))))))....)))))..(((((......))))))))) ( -35.40, z-score = -1.31, R) >droAna3.scaffold_13340 5065073 103 + 23697760 ------------ACGUGAUUCGCC------UAGAAUGAUGUCCAGCAUGAGCAUGGGUGACUUUAUUUCGCCCACGGGCUCCAAUCUAAGUCCCUUGGAGAACUCCGCUCUGAGAGUUUCC ------------((((.((((...------..)))).))))....((.((((.(((((((.......))))))).(..((((((..........))))))..)...))))))......... ( -30.60, z-score = -1.03, R) >dp4.chr2 4380249 106 + 30794189 ------------GUAUCACUUUCU---UUAUAGAAUGAUGUCUAGCAUGAGCAUGGGUGACUUCAUCUCGCCCACUGGUUCGAAUCUGAGUCCAUUGGAGAACUCCGCUCUGCGUGUCUCA ------------.(((((..((((---....)))))))))....(((.((((.(((((((.......))))))).(((((((....)))).)))..(((....))))))))))........ ( -30.50, z-score = -1.17, R) >droPer1.super_19 84077 106 + 1869541 ------------GUCUUACUUUCU---UUAUAGAAUGAUGUCUAGCAUGAGCAUGGGUGACUUCAUCUCGCCCACUGGUUCGAAUCUGAGUCCAUUGGAGAACUCCGCUCUGCGUGUCUCA ------------(...(((.....---...((((......))))(((.((((.(((((((.......))))))).(((((((....)))).)))..(((....)))))))))))))...). ( -28.60, z-score = -0.70, R) >droWil1.scaffold_181108 3223157 110 + 4707319 ----------AUAUGCAUUUUUCG-UUUGCCAGAAUGAUGUCAAGCAUGAGUAUGGGUGAUUUUAUCUCACCCACUGGCUCAAAUCUGAGUCCAUUGGAGAACUCAGCGUUACGUGUAUCG ----------..((((((...(((-(((....)))))).((...((.(((((.(((((((.......))))))).(((((((....)))).))).......)))))))...)))))))).. ( -31.60, z-score = -1.82, R) >droVir3.scaffold_13047 12571413 120 + 19223366 UGUCCAACUCUCCCAUGUUUUAUC-UUCUGCAGCAUGAUGUCCAGCAUGAGCAUGGGCGACUUUAUCUCACCUACGGGCUCCAAUCUGAGUCCCUUGGAGAAUUCAGCGUUACGCGUUUCG ..(((((.((.(((((((((...(-(...(((......)))..))...))))))))).))...............((((((......)))))).)))))(((....(((...)))..))). ( -31.90, z-score = -0.88, R) >droGri2.scaffold_14830 5736870 119 - 6267026 -UUCUACAUCUUUCAUCUCUUAUU-AUUCCCAGCAUGAUGUCUAGCAUGAGCAUGGGCGAUUUUAUCUCACCCACUGGCUCCAAUCUAAGUCCAUUGGAGAACUCAGCAUUGCGCGUUUCC -.......................-.......((..(((((......((((((((((.((.......)).)))).)).(((((((........)))))))..)))))))))..))...... ( -23.50, z-score = -0.03, R) >droMoj3.scaffold_6540 4894307 118 - 34148556 --UCCAUAUCUCGUUUCUUUGAUC-GCUCGCAGCAUGAUGUCUAGCAUGAGCAUGGGUGACUUUAUCUCGCCCACCGGCUCCAAUCUGAGCCCCUUGGAGAACUCGACGCUGCGCGUUUCG --..................((..-((.((((((...(((.....)))(((..(((((((.......)))))))(((((((......)))))....))....)))...)))))).)).)). ( -34.60, z-score = -0.69, R) >anoGam1.chr2R 55674858 114 - 62725911 -------ACAAGGAUCUUCUCGUUUGCUUGCAGAAUGAUGUCCAGCAUGAGCAUGGGUGACUUCAUCUCGCCAACUGGAUCCAGCUUGAGUCCGCUCGACAGCUCGGGACUGCGGGUGUCG -------..................((((((((......((((((..((.((..(((((....))))).)))).))))))((((((((((....))))..)))).))..)))))))).... ( -34.60, z-score = 0.92, R) >consensus ____________AUGUGUAUUUCC_AACUGCAGAAUGAUGUCCAGCAUGAGCAUGGGUGACUUUAUCUCGCCCACUGGCUCCAAUCUGAGUCCUUUGGAGAACUCGGCUCUGCGCGUGUCC ..............................((((..............((((.(((((((.......)))))))...))))....((((((.(......).)))))).))))......... (-18.72 = -18.88 + 0.16)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:59:28 2011