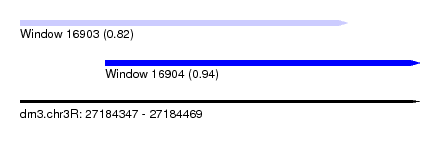
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 27,184,347 – 27,184,469 |
| Length | 122 |
| Max. P | 0.943082 |

| Location | 27,184,347 – 27,184,447 |
|---|---|
| Length | 100 |
| Sequences | 11 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 78.42 |
| Shannon entropy | 0.43327 |
| G+C content | 0.48443 |
| Mean single sequence MFE | -28.16 |
| Consensus MFE | -19.19 |
| Energy contribution | -18.72 |
| Covariance contribution | -0.47 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.818437 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
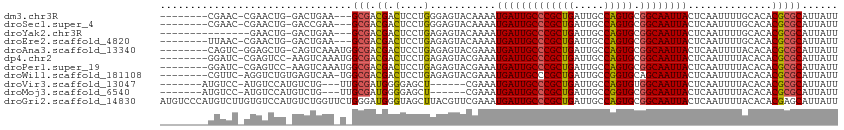
>dm3.chr3R 27184347 100 + 27905053 --------CGAAC-CGAACUG-GACUGAA---GCGACGACUCCUGGGAGUACAAAAUGAUUGCCCGCUGAUUGCCAGUGCGGCAAUUACUCAAUUUUGCACACGCGCAUUAUU --------....(-(.....)-)......---(((.((((((....)))).((((((((((((((((((.....))))).))))))).....))))))....)))))...... ( -29.10, z-score = -2.26, R) >droSec1.super_4 5989545 100 + 6179234 --------CGAAC-CGAACUG-GACCGAA---GCGACGACUCCUGGGAGUACAAAAUGAUUGCCCGCUGAUUGCCAGUGCGGCAAUUACUCAAUUUUGCACACGCGCAUUAUU --------((..(-(.....)-)..))..---(((.((((((....)))).((((((((((((((((((.....))))).))))))).....))))))....)))))...... ( -29.80, z-score = -2.37, R) >droYak2.chr3R 28113753 94 + 28832112 ---------------GAACUG-GACUGAA---GCGACGACUCCUGAGAGUACAAAAUGAUUGCCCGCUGAUUGCCAGUGCGGCAAUUACUCAAUUUUGCACACGCGCAUUAUU ---------------......-.......---(((.((((((....)))).((((((((((((((((((.....))))).))))))).....))))))....)))))...... ( -26.80, z-score = -2.38, R) >droEre2.scaffold_4820 9668979 100 - 10470090 --------UUAAC-CGAACUG-GACUGAA---GCGACGACUCCUGAGAGUACAAAAUGAUUGCCCGCUGAUUGCCAGUGCGGCAAUUACUCAAUUUUGCACACGCGCAUUAUU --------....(-(.....)-)......---(((.((((((....)))).((((((((((((((((((.....))))).))))))).....))))))....)))))...... ( -27.20, z-score = -2.60, R) >droAna3.scaffold_13340 5062750 103 + 23697760 --------CAGUC-GGAGCUG-CAGUCAAAUGGCGACGACUCCUGAGAGUACGAAAUGAUUGCCCGCUGAUUGCCAGUGCGGCAAUUACUCAAUUUUACACACGCGCAUUAUU --------((((.-...))))-......(((((((.((((((....)))).))...(((((((((((((.....))))).))))))))..............))).))))... ( -30.20, z-score = -1.25, R) >dp4.chr2 4378365 103 + 30794189 --------GGAUC-CGAGUCC-AAGUCAAAUGGCGACGACUCCUGAGAGUACGAAAUGAUUGCCCGCUGAUUGCCAGUGCGGCAAUUACUCAAUUUUACACACGCGCAUUAUU --------((((.-...))))-......(((((((.((((((....)))).))...(((((((((((((.....))))).))))))))..............))).))))... ( -31.90, z-score = -2.67, R) >droPer1.super_19 82190 103 + 1869541 --------GGAUC-CGAGUCC-AAGUCAAAUGGCGACGACUCCUGAGAGUACGAAAUGAUUGCCCGCUGAUUGCCAGUGCGGCAAUUACUCAAUUUUACACACGCGCAUUAUU --------((((.-...))))-......(((((((.((((((....)))).))...(((((((((((((.....))))).))))))))..............))).))))... ( -31.90, z-score = -2.67, R) >droWil1.scaffold_181108 3221282 103 + 4707319 --------CGUUC-AGGUCUGUGAGUCAA-UGGCGACGACUCCUGAGAGUACGAAAUGAUUGCCCGCUGAUUGCCGGUGCAGCAAUUACUCAAUUUUACACACGCGCAUUAUU --------(((((-(((...((..(((..-....))).)).)))))(((((.......(((((.(((((.....)))))..))))))))))............)))....... ( -23.91, z-score = 0.72, R) >droVir3.scaffold_13047 12569563 96 + 19223366 -------AUGUCC-AUGUCCAUGUCUG---UUGCGAUGGGGAGCU------CGAAAUGAUUGCCCGCUGAUUGCCAGUGUGGCAAUUACUCAAUUUUACACACGCGCAUUAUU -------((((.(-.(((((..(((..---....)))..)))...------.((..(((((((((((((.....))))).)))))))).)).........)).).)))).... ( -23.60, z-score = 0.57, R) >droMoj3.scaffold_6540 4892439 96 - 34148556 -------AUGUCC-AUGUCCAUGUCUG---UUGCGAUGGGGAGCU------CGAAAUGAUUGCCCGCUGAUUGCCGGUGCGGCAAUUACUCAAUUUUACACACGCGCAUUAUU -------((((.(-.(((((..(((..---....)))..)))...------.((..(((((((((((((.....))))).)))))))).)).........)).).)))).... ( -22.90, z-score = 1.27, R) >droGri2.scaffold_14830 5734362 113 - 6267026 AUGUCCCAUGUCUUGUGUCCAUGUCUGGUUCUGGGAUGGGUAGCUUACGUUCGAAAUGAUUGCCCGCUGAUUGCCAGUGCGGCAAUUACUCAAUUUUACACACGAGCAUUAUU .......(((.(((((((((((.((.......)).)))).............((..(((((((((((((.....))))).)))))))).))........)))))))))).... ( -32.40, z-score = -0.95, R) >consensus ________CGAUC_CGAACUG_GACUGAA_U_GCGACGACUCCUGAGAGUACGAAAUGAUUGCCCGCUGAUUGCCAGUGCGGCAAUUACUCAAUUUUACACACGCGCAUUAUU ................................(((.((.(....)...........(((((((((((((.....))))).))))))))..............)))))...... (-19.19 = -18.72 + -0.47)
| Location | 27,184,373 – 27,184,469 |
|---|---|
| Length | 96 |
| Sequences | 11 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 84.25 |
| Shannon entropy | 0.32912 |
| G+C content | 0.47944 |
| Mean single sequence MFE | -29.55 |
| Consensus MFE | -24.18 |
| Energy contribution | -24.37 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.943082 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
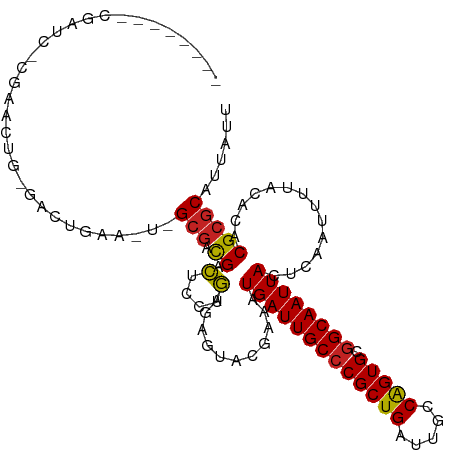
>dm3.chr3R 27184373 96 + 27905053 -CUCCUGGGAGUACAAAAUGAUUGCCCGCUGAUUGCCAGUGCGGCAAUUACUCAAUUUUGCACACGCGCAUUAUUUGUCUGUGUCGAUCUCGUGGUG----------- -..(((((((........(((((((((((((.....))))).)))))))).......(((.(((((.(((.....))).))))))))))))).))..----------- ( -30.50, z-score = -1.89, R) >droSec1.super_4 5989571 96 + 6179234 -CUCCUGGGAGUACAAAAUGAUUGCCCGCUGAUUGCCAGUGCGGCAAUUACUCAAUUUUGCACACGCGCAUUAUUUGUCUGUGUCGAUCUCGUGGUG----------- -..(((((((........(((((((((((((.....))))).)))))))).......(((.(((((.(((.....))).))))))))))))).))..----------- ( -30.50, z-score = -1.89, R) >droYak2.chr3R 28113773 107 + 28832112 -CUCCUGAGAGUACAAAAUGAUUGCCCGCUGAUUGCCAGUGCGGCAAUUACUCAAUUUUGCACACGCGCAUUAUUUGUCUGUGUCGAUCUCGUGGCAUGGUUACAUGG -..(((((((........(((((((((((((.....))))).)))))))).......(((.(((((.(((.....))).))))))))))))).))((((....)))). ( -34.80, z-score = -2.05, R) >droEre2.scaffold_4820 9669005 100 - 10470090 -CUCCUGAGAGUACAAAAUGAUUGCCCGCUGAUUGCCAGUGCGGCAAUUACUCAAUUUUGCACACGCGCAUUAUUUGUCUGUGUCGAUCUCGUGGCAUGGU------- -..(((((((........(((((((((((((.....))))).)))))))).......(((.(((((.(((.....))).))))))))))))).))......------- ( -31.20, z-score = -1.50, R) >droAna3.scaffold_13340 5062779 90 + 23697760 -CUCCUGAGAGUACGAAAUGAUUGCCCGCUGAUUGCCAGUGCGGCAAUUACUCAAUUUUACACACGCGCAUUAUUUGUCUGUGUCGAUCUC----------------- -.....((((...(((..(((((((((((((.....))))).))))))))............((((.(((.....))).))))))).))))----------------- ( -27.40, z-score = -2.86, R) >dp4.chr2 4378394 103 + 30794189 -CUCCUGAGAGUACGAAAUGAUUGCCCGCUGAUUGCCAGUGCGGCAAUUACUCAAUUUUACACACGCGCAUUAUUUGUCUGUGUCGAUCUUG-GAUGUGGAGGUG--- -((((...(((..(((..(((((((((((((.....))))).))))))))............((((.(((.....))).)))))))..))).-.....))))...--- ( -31.00, z-score = -1.49, R) >droPer1.super_19 82219 103 + 1869541 -CUCCUGAGAGUACGAAAUGAUUGCCCGCUGAUUGCCAGUGCGGCAAUUACUCAAUUUUACACACGCGCAUUAUUUGUCUGUGUCGAUCUUG-GAUGUGGAGAUG--- -((((...(((..(((..(((((((((((((.....))))).))))))))............((((.(((.....))).)))))))..))).-.....))))...--- ( -30.80, z-score = -1.84, R) >droWil1.scaffold_181108 3221311 102 + 4707319 -CUCCUGAGAGUACGAAAUGAUUGCCCGCUGAUUGCCGGUGCAGCAAUUACUCAAUUUUACACACGCGCAUUAUUUGUCUGUGUCGAUCUGUGUUCGAGGGCG----- -..(((..((((((((..(((((((.(((((.....)))))..))))))).))........(((((.(((.....))).)))))......))))))..)))..----- ( -25.60, z-score = 0.26, R) >droVir3.scaffold_13047 12569590 87 + 19223366 ------GGGAGCUCGAAAUGAUUGCCCGCUGAUUGCCAGUGUGGCAAUUACUCAAUUUUACACACGCGCAUUAUUUGUCUGUGUCGAUCUCGC--------------- ------..(((.((((..(((((((((((((.....))))).))))))))............((((.(((.....))).)))))))).)))..--------------- ( -30.30, z-score = -2.97, R) >droMoj3.scaffold_6540 4892466 87 - 34148556 ------GGGAGCUCGAAAUGAUUGCCCGCUGAUUGCCGGUGCGGCAAUUACUCAAUUUUACACACGCGCAUUAUUUGUCUGUGUCGAUCUGAC--------------- ------...((.((((..(((((((((((((.....))))).))))))))............((((.(((.....))).)))))))).))...--------------- ( -26.10, z-score = -1.44, R) >droGri2.scaffold_14830 5734400 93 - 6267026 GGUAGCUUACGUUCGAAAUGAUUGCCCGCUGAUUGCCAGUGCGGCAAUUACUCAAUUUUACACACGAGCAUUAUUUGUCUGUGUCGAUCUCGC--------------- .........((.((((..(((((((((((((.....))))).))))))))............((((..((.....))..))))))))...)).--------------- ( -26.90, z-score = -1.82, R) >consensus _CUCCUGAGAGUACGAAAUGAUUGCCCGCUGAUUGCCAGUGCGGCAAUUACUCAAUUUUACACACGCGCAUUAUUUGUCUGUGUCGAUCUCGUG__G___________ ......((((........(((((((((((((.....))))).))))))))...........(((((.(((.....))).)))))...))))................. (-24.18 = -24.37 + 0.19)

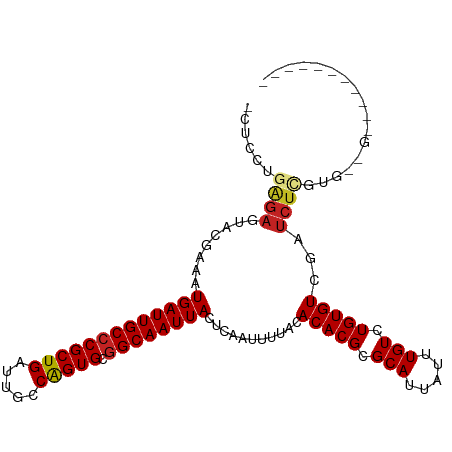
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:59:26 2011