| Sequence ID | dm3.chr3R |
|---|---|
| Location | 27,149,833 – 27,149,978 |
| Length | 145 |
| Max. P | 0.691246 |

| Location | 27,149,833 – 27,149,978 |
|---|---|
| Length | 145 |
| Sequences | 6 |
| Columns | 153 |
| Reading direction | forward |
| Mean pairwise identity | 58.55 |
| Shannon entropy | 0.77630 |
| G+C content | 0.41093 |
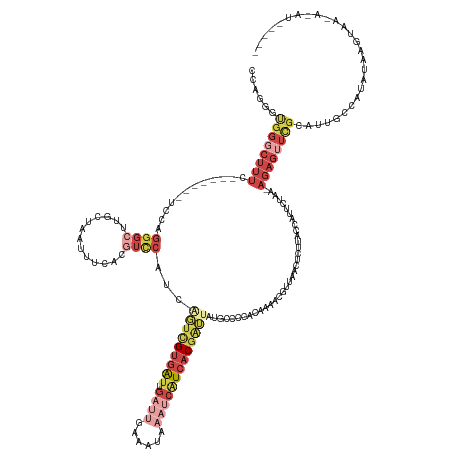
| Mean single sequence MFE | -33.97 |
| Consensus MFE | -6.82 |
| Energy contribution | -10.44 |
| Covariance contribution | 3.62 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.20 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.691246 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
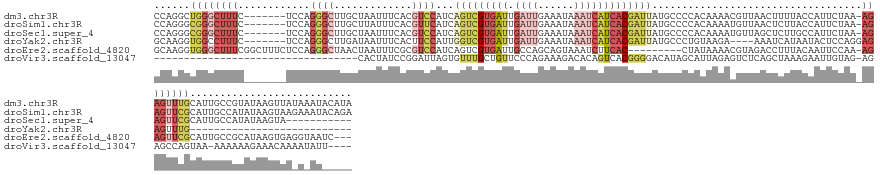
>dm3.chr3R 27149833 145 + 27905053 CCAGGCUGGGCUUUC-------UCCAGGGCUUGCUAAUUUCACGUCCAUCAGUCGUGAUUGAUUGAAAUAAAUCAUCACGAUUAUGCCCCACAAAACGUUAACUUUUACCAUUCUAA-AGAGUUUGCAUUGCCGUAUAAGUUAUAAAUACAUA ...(((.((((....-------....((((.((.......)).))))...(((((((((.((((......)))))))))))))..))))........((.(((((((.........)-)))))).))...)))((((.........))))... ( -36.80, z-score = -2.80, R) >droSim1.chr3R 26789392 145 + 27517382 CCAGGGCGGGCUUUC-------UCCAGGGCUUGCUUAUUUCACGUUCAUCAGUCGUGAUUGAUUGAAAUAAAUCAUCACGAUUAUGCCCCACAAAAUGUUAACUCUUACCAUUCUAA-AGAGUUCGCAUUGCCAUAUAAGUAAGAAAUACAGA ....(((((((((..-------....)))))))))((((((..(..(((.(((((((((.((((......))))))))))))))))..).....(((((.(((((((.........)-)))))).))))).............)))))).... ( -40.00, z-score = -3.13, R) >droSec1.super_4 5960185 134 + 6179234 CCAGGGCGGGCUUUC-------UCCAGGGCUUGCUAAUUUCACGUCCAUCAGUCGUGAUUGAUUGAAAUAAAUCAUCACGAUUAUGCCCCACAAAAUGUUAGCUCUUGCCAUUCUAA-AGAGUUCGCAUUGCCAUAUAAGUA----------- ....(((((((....-------....((((.((.......)).))))...(((((((((.((((......)))))))))))))..)))).....(((((.(((((((.........)-)))))).)))))))).........----------- ( -42.80, z-score = -3.88, R) >droYak2.chr3R 28083353 115 + 28832112 GCAAGGUGGCCUUUC-------UCCAGGGCUUGAUAAUUUCACUUCCAUUGGUCGUGAUUGAUUGAAAUAAAUCAUCACGAUUAUGCCCUGUAAGA----AAAUCAUAAUACUCCAGGAGAGUUUG--------------------------- .....((((..((((-------(.((((((.(((.....))).......((((((((((.((((......)))))))))))))).))))))..)))----)).))))...((((.....))))...--------------------------- ( -32.80, z-score = -2.36, R) >droEre2.scaffold_4820 9639584 140 - 10470090 GCAAGGUGGGCUUUCGGCUUUCUCCAGGGCUAACUAAUUUCGCGUCCAUCAGUCGUGAUUGCCAGCAGUAAAUCUUCAC---------CUAUAAAACGUAGACCUUUACAAUUCCAA-AGAGUUCGCAUUGCCGCAUAAGUGAGGUAAUC--- ...((((((((....(((((......)))))........(((((.(.....).)))))..)))...((.....)).)))---------)).......(..(((((((........))-)).)))..)((((((.((....)).)))))).--- ( -29.80, z-score = 0.54, R) >droVir3.scaffold_13047 12537198 113 + 19223366 ----------------------------------CACUAUCCGGAUUAGUGUUUGCUGUUCCCAGAAAGACACAGUCACGGGGACAUAGCAUUAGAGUCUCAGCUAAAGAAUUGUAG-AGAGCCAGUAA-AAAAAAGAAACAAAAUAUU---- ----------------------------------........((.((((((((...(((((((.....(((...)))..))))))).))))))))..((((.((.........)).)-))).)).....-...................---- ( -21.60, z-score = -0.78, R) >consensus CCAGGGUGGGCUUUC_______UCCAGGGCUUGCUAAUUUCACGUCCAUCAGUCGUGAUUGAUUGAAAUAAAUCAUCACGAUUAUGCCCCACAAAACGUUAACUCUUACCAUUCUAA_AGAGUUCGCAUUGCCAUAUAAGUAA_A_AU_____ ......((((((((............((((.............))))...(((((((((.((((......)))))))))))))...................................))))))))........................... ( -6.82 = -10.44 + 3.62)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:59:20 2011