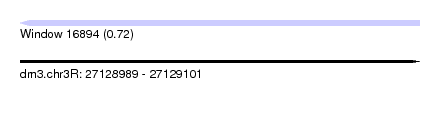
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 27,128,989 – 27,129,101 |
| Length | 112 |
| Max. P | 0.722421 |

| Location | 27,128,989 – 27,129,101 |
|---|---|
| Length | 112 |
| Sequences | 15 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.14 |
| Shannon entropy | 0.50043 |
| G+C content | 0.50000 |
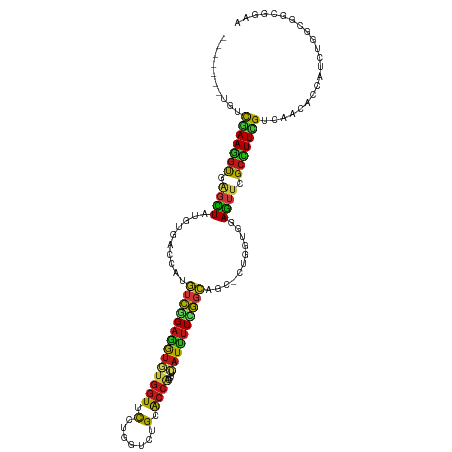
| Mean single sequence MFE | -37.78 |
| Consensus MFE | -19.17 |
| Energy contribution | -18.31 |
| Covariance contribution | -0.85 |
| Combinations/Pair | 1.77 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.722421 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
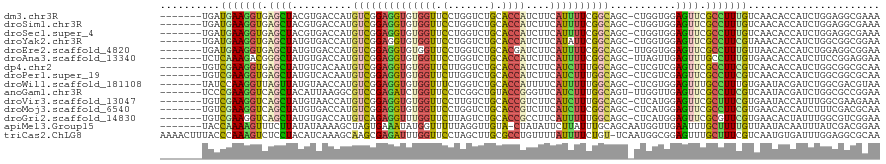
>dm3.chr3R 27128989 112 - 27905053 -------UGAUGAAGGUGAGCUACGUGACCAUGUCGGAGGUGUGGUUCCUGGUCUGCACCAUCUUCAUUUUCGGCAGC-CUGGUGGAGUUCGCCUUUGUCAACACCAUCUGGAGGCGAAA -------(((..((((((((((...(.(((((((((((((((.((....((((....)))).)).)))))))))))..-.)))).)))))))))))..)))...((....))........ ( -41.50, z-score = -1.31, R) >droSim1.chr3R 26768207 112 - 27517382 -------UGAUGAAGGUGAGCUACGUGACCAUGUCGGAGGUGUGGUUCCUGGUCUGCACCAUCUUCAUUUUCGGCAGC-CUGGUGGAGUUCGCCUUUGUCAACACCAUCUGGAGGCGAAA -------(((..((((((((((...(.(((((((((((((((.((....((((....)))).)).)))))))))))..-.)))).)))))))))))..)))...((....))........ ( -41.50, z-score = -1.31, R) >droSec1.super_4 5939506 112 - 6179234 -------UGAUGAAGGUGAGCUACGUGACCAUGUCGGAGGUGUGGUUCCUGGUCUGCACCAUCUUCAUUUUCGGCAGC-CUGGUGGAGUUCGCCUUUGUCAACACCAUCUGGAGGCGAAA -------(((..((((((((((...(.(((((((((((((((.((....((((....)))).)).)))))))))))..-.)))).)))))))))))..)))...((....))........ ( -41.50, z-score = -1.31, R) >droYak2.chr3R 28061266 112 - 28832112 -------UGAUGAAGGUGAGCUAUGUGACCAUGUCGGAGGUGUGGUUCCUGGUCUGCACCAUCUUCAUAUUCGGCAGC-CUGGUGGAGUUCGCCUUCGUAAACACCAUCUGGCGGCGGAA -------(((.((.(((((((((.(..((((..(.....)..))))..)))).)).)))).)).)))..(((.((.((-(.(((((.(((..(....)..))).))))).))).)).))) ( -42.40, z-score = -1.44, R) >droEre2.scaffold_4820 9618134 112 + 10470090 -------UGAUGAAGGUGAGCUAUGUGACCAUGUCGGAGGUGUGGUUCCUGGUCUGCACGAUCUUCAUUUUCGGCAGC-UUGGUGGAGUUCGCCUUUGUUAACACCAUCUGGAGGCGGAA -------(((..((((((((((...(.(((((((((((((((.((.((.((.....)).)).)).)))))))))))..-.)))).)))))))))))..)))......((((....)))). ( -39.90, z-score = -1.38, R) >droAna3.scaffold_13340 5008736 112 - 23697760 -------UCUCAAAGACGGGCUAUGUGACCAUGUCGGAGGUGUGGUUCCUGGUCUGCACCAUCUUCAUUUUCGGCAGC-UUAGUUGAGUUUGCCUUUGUGAACACCAUCUUCCGGAGGAA -------((((...(((((((.....).)).))))(((((.(((((...((((....))))..(((((....((((((-((....))).)))))...))))).)))))))))).)))).. ( -34.20, z-score = -0.34, R) >dp4.chr2 4325106 112 - 30794189 -------UGUCGAAGGUGAGCUAUGUCACAAUGUCGGAGGUGUGGUUCUUGGUCUGCACCAUCUUCAUCUUUGGCAGC-CUCGUCGAGUUCGCCUUCGUCAACACCAUCUGGCGGCGCAA -------.((((((((((((((.........(((((((((((.((....((((....)))).)).)))))))))))..-.......)))))))))))((((........))))))).... ( -42.87, z-score = -2.60, R) >droPer1.super_19 29007 112 - 1869541 -------UGUCGAAGGUGAGCUAUGUCACAAUGUCGGAGGUGUGGUUCUUGGUCUGCACCAUCUUCAUCUUUGGCAGC-CUCGUCGAGUUCGCCUUCGUCAACACCAUCUGGCGGCGCAA -------.((((((((((((((.........(((((((((((.((....((((....)))).)).)))))))))))..-.......)))))))))))((((........))))))).... ( -42.87, z-score = -2.60, R) >droWil1.scaffold_181108 3163277 112 - 4707319 -------UAUCCAAGGUUAGUUAUGUAACCAUGUCGGAGGUGUGGUUUCUGGUCUGCACCAUUUUCAUUUUUGGCAGC-CUCGUGGAGUUUGCCUUUGUGAAUACGAUCUGGCGACGUAA -------.............((((((..(((.((((..((((..(........)..))))...(((((....(((((.-(((...))).)))))...)))))..)))).)))..)))))) ( -35.20, z-score = -1.92, R) >anoGam1.chr3R 15541487 112 + 53272125 -------UCCCGAAGGUCAGCUACAUUAAGGCGUCCGAGAUCUGGUUCCUCGGCUGUACCGGGUUCAUCUUUGGCAGU-UUGGUUGAGUUCGCCUUCGUCAAUACGAUCUGGCGCCGGAA -------(((.((((((.((((.((...(((((((.(((((...(..((.(((.....)))))..)))))).))).))-))...)))))).))))))((((........))))...))). ( -34.10, z-score = 0.31, R) >droVir3.scaffold_13047 12512611 112 - 19223366 -------UGUCGAAGGUCAGCUAUGUAACCAUGUCGGAGGUGUGGUUCCUUGUCUGCACCGUCUUCAUCUUUGGCAGC-CUCAUGGAGUUCGCUUUCGUGAAUACCAUUUGGCGAAGAAA -------((((((((((..(.((((....)))).)(..((((..(........)..))))..)...))))))))))((-(..((((.((((((....)))))).))))..)))....... ( -38.60, z-score = -2.39, R) >droMoj3.scaffold_6540 4836866 112 + 34148556 -------UGUCGAAGGUCAGCUAUGUGACCAUGUCGGAGGUGUGGUUCCUGGUCUGCACCGUCUUCAUCUUCGGCAGC-CUCAUGGAGUUCGCCUUCGUGAACACCAUCUUUCGACGCAA -------.(((((((((((......))))).(((((((((((.((....((((....)))).)).)))))))))))..-...((((.((((((....)))))).))))..)))))).... ( -43.00, z-score = -2.52, R) >droGri2.scaffold_14830 5660076 112 + 6267026 -------UGUCGAAGGUCAGCUAUGUGACCAUGUCAGAGGUUUGGUUCUUAGUCUGCACCGCCUUCAUUUUUGGCAGC-CUCAUGGAGUUCGCGUUCGUGAACACUAUUUGGCGUCGGAA -------....((((((((((((...(((((..(.....)..)))))..))).)))....))))))..(((((((.((-(..((((.((((((....)))))).))))..)))))))))) ( -38.70, z-score = -2.05, R) >apiMel3.Group15 3274 112 - 7856270 -------UACCAAAAGUUUCUUAUAUAAAAGCUAGUGAAAUAUGGUUUUUAGGUUGUA-CUAUAUUCUUAUUUGCAGCAAUGGUUGAAUUUGCUUUUGUUAAUACAAUUUAUCGACGGAA -------..(((((((((((.....(((((((((........))))))))).((((((-..(((....))).)))))).......)))...))))))((..(((.....)))..)))).. ( -19.00, z-score = -0.21, R) >triCas2.ChLG8 7606183 119 + 15773733 AAAACUUUACCCAAAGUCUCCUACAUCAAAGCAAGCGAGAUUUGGUUCCUAGCUUGCGCCUGUUUUAUUUUCUGU-UCAAUGGCGGAGUUUGCUUUCGUCAAUGUGAUUUGGAGGCGCAA ...............((((((...((((..((((((..((......))...))))))..................-.((.((((((((.....)))))))).))))))..)))))).... ( -31.30, z-score = -0.85, R) >consensus _______UGUCGAAGGUGAGCUAUGUGACCAUGUCGGAGGUGUGGUUCCUGGUCUGCACCAUCUUCAUUUUCGGCAGC_CUGGUGGAGUUCGCCUUCGUCAACACCAUCUGGCGGCGGAA ..........(((((((.((((..........((((((((((((((.(.......).))))....))))))))))...........)))).)))))))...................... (-19.17 = -18.31 + -0.85)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:59:18 2011