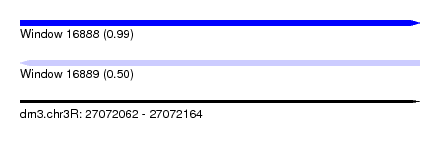
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 27,072,062 – 27,072,164 |
| Length | 102 |
| Max. P | 0.992024 |

| Location | 27,072,062 – 27,072,164 |
|---|---|
| Length | 102 |
| Sequences | 9 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 69.62 |
| Shannon entropy | 0.61278 |
| G+C content | 0.43199 |
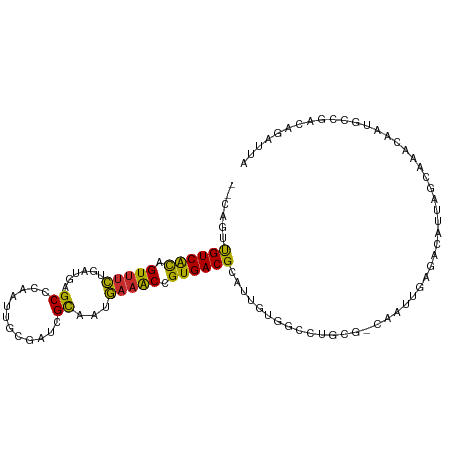
| Mean single sequence MFE | -27.34 |
| Consensus MFE | -13.01 |
| Energy contribution | -13.90 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.51 |
| SVM RNA-class probability | 0.992024 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
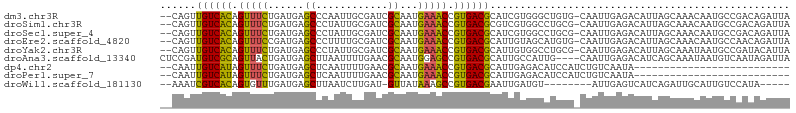
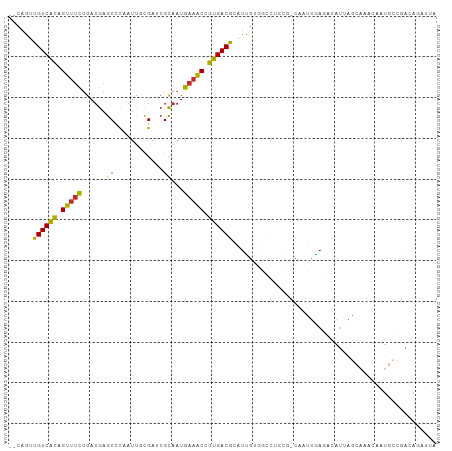
>dm3.chr3R 27072062 102 + 27905053 UAAUCUGUCGGCAUUGUUUGCUAAUGUCUCAAUUG-CACAGCCCACGAUGCGUCACGGUUUCAUUGCGAUCGCAAUUGGGCUCAUCAGAAACUGUGACAACUG-- ..........(((((((..(((..(((.......)-)).)))..)))))))((((((((((((((((....)))))(((.....)))))))))))))).....-- ( -32.80, z-score = -2.46, R) >droSim1.chr3R 26715171 102 + 27517382 UAAUCUGUCGGCAUUGUUUGCUAAUGUCUCAAUUG-CGCAGGCCACGACGCGUCACGGUUUCAUUGCGAUCGCAAUAGGGCUCAUCAGAAACUGUGACAACUG-- ......((((.....((((((((((......))))-.))))))..))))..((((((((((((((((....))))).((.....)).))))))))))).....-- ( -33.70, z-score = -2.30, R) >droSec1.super_4 5889723 102 + 6179234 UAAUCUGUCGGCAUUGUUUGCUAAUGUCUCAAUUG-CGCAGGCCACGAUGCGUCACGGUUUCAUUGCGAUCGCAAUAGGGCUCAUCAGAAACUGUGACAACUG-- ..........(((((((((((((((......))))-.))))...)))))))((((((((((((((((....))))).((.....)).))))))))))).....-- ( -33.60, z-score = -2.27, R) >droEre2.scaffold_4820 9562527 102 - 10470090 UAAUCUGUUGGCAUUGUUUGCUAAUGUCUCAAUUG-CACAUGCUACAAUGCGUCACGGUUUCAUUGCGAUCGCAAAAGGGCUCAUCGGAAACUGUGACAACUG-- ......(((.(((((((..((..(((((......)-.)))))).)))))))(((((((((((.((((....))))..((.....)).))))))))))))))..-- ( -32.40, z-score = -3.03, R) >droYak2.chr3R 28008681 102 + 28832112 UAAUGUAUCGGCAUUAUUUGCUAAUGUCUCAAUUG-CGCAGGCCACAAUGCGUCACGGUUUCAUUGCGAUCGCAAUAGGGCUCAUCAGAAACUGUGACAACUG-- .........(((((((.....))))))).......-.(((........)))((((((((((((((((....))))).((.....)).))))))))))).....-- ( -29.80, z-score = -1.84, R) >droAna3.scaffold_13340 4955109 101 + 23697760 UAAUCUAUUGACAUUAUUUGCUGAUGUCUCAAUUG----CAAUGGCAAUGCGUCACGGCUCCAUUGCGUUCAAAAUUAAGCUCAUCAGUAACUGCGACAUCGGAG ....................((((((((.((.(((----(....((((((.((....))..))))))(((........)))......)))).)).)))))))).. ( -24.30, z-score = -1.07, R) >dp4.chr2 4266079 77 + 30794189 --------------------------UAUUGACAGAUGGAUGUCUCAAUGCGUCACGGUUUCAUUGCGUUCAAAAUUGAGCUCAUCAGAAACUAUGACAAUUG-- --------------------------....((((......)))).((((..((((..(((((..((.(((((....))))).))...)))))..)))).))))-- ( -22.50, z-score = -2.58, R) >droPer1.super_7 4359139 77 - 4445127 --------------------------UAUUGACAGAUGGAUGUCUCAAUGCGUCACGGUUUCAUUGCGUUCAAAAUUGAGCUCAUCAGAAACUAUGACAAUUG-- --------------------------....((((......)))).((((..((((..(((((..((.(((((....))))).))...)))))..)))).))))-- ( -22.50, z-score = -2.58, R) >droWil1.scaffold_181130 10494467 89 + 16660200 -----UAUGGACAAUGCAAUCUGAUGACUCAAU--------ACAUCAAUUCGUCACGGCUUUAUAAC-AUCAAGAUUAAGCUCAUCAAACACUGUGACGAUUU-- -----................(((((.......--------.)))))..((((((((((((......-.........)))))...........)))))))...-- ( -14.46, z-score = -0.73, R) >consensus UAAUCUGUCGGCAUUGUUUGCUAAUGUCUCAAUUG_CGCAGGCCACAAUGCGUCACGGUUUCAUUGCGAUCGCAAUUGGGCUCAUCAGAAACUGUGACAACUG__ ...................................................(((((((((((..((.(.((......)).).))...)))))))))))....... (-13.01 = -13.90 + 0.89)



| Location | 27,072,062 – 27,072,164 |
|---|---|
| Length | 102 |
| Sequences | 9 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 69.62 |
| Shannon entropy | 0.61278 |
| G+C content | 0.43199 |
| Mean single sequence MFE | -24.10 |
| Consensus MFE | -10.50 |
| Energy contribution | -9.89 |
| Covariance contribution | -0.61 |
| Combinations/Pair | 1.46 |
| Mean z-score | -1.01 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>dm3.chr3R 27072062 102 - 27905053 --CAGUUGUCACAGUUUCUGAUGAGCCCAAUUGCGAUCGCAAUGAAACCGUGACGCAUCGUGGGCUGUG-CAAUUGAGACAUUAGCAAACAAUGCCGACAGAUUA --..((((.((..((((((((((((((((..((((.((((.........))))))))...))))))...-(....)...)))))).))))..)).))))...... ( -32.00, z-score = -1.91, R) >droSim1.chr3R 26715171 102 - 27517382 --CAGUUGUCACAGUUUCUGAUGAGCCCUAUUGCGAUCGCAAUGAAACCGUGACGCGUCGUGGCCUGCG-CAAUUGAGACAUUAGCAAACAAUGCCGACAGAUUA --..((((.((..((((((((((..(.(.((((((...((.((((...((.....)))))).))...))-)))).).).)))))).))))..)).))))...... ( -25.70, z-score = 0.04, R) >droSec1.super_4 5889723 102 - 6179234 --CAGUUGUCACAGUUUCUGAUGAGCCCUAUUGCGAUCGCAAUGAAACCGUGACGCAUCGUGGCCUGCG-CAAUUGAGACAUUAGCAAACAAUGCCGACAGAUUA --..((((.((..((((((((((.(((....((((.((((.........))))))))....)))((.((-....)))).)))))).))))..)).))))...... ( -25.10, z-score = -0.07, R) >droEre2.scaffold_4820 9562527 102 + 10470090 --CAGUUGUCACAGUUUCCGAUGAGCCCUUUUGCGAUCGCAAUGAAACCGUGACGCAUUGUAGCAUGUG-CAAUUGAGACAUUAGCAAACAAUGCCAACAGAUUA --..((((((((.(((((((((..((......)).))))....))))).)))))(((((((.((..(((-(......).)))..))..))))))).)))...... ( -27.10, z-score = -1.76, R) >droYak2.chr3R 28008681 102 - 28832112 --CAGUUGUCACAGUUUCUGAUGAGCCCUAUUGCGAUCGCAAUGAAACCGUGACGCAUUGUGGCCUGCG-CAAUUGAGACAUUAGCAAAUAAUGCCGAUACAUUA --((((((.(.(((..((....))(((.((.((((.((((.........)))))))).)).)))))).)-))))))....((..(((.....)))..))...... ( -24.80, z-score = -0.40, R) >droAna3.scaffold_13340 4955109 101 - 23697760 CUCCGAUGUCGCAGUUACUGAUGAGCUUAAUUUUGAACGCAAUGGAGCCGUGACGCAUUGCCAUUG----CAAUUGAGACAUCAGCAAAUAAUGUCAAUAGAUUA ....(((((.....((.((((((..(((((((......(((((((.((......))....))))))----)))))))).)))))).))...)))))......... ( -23.30, z-score = -0.32, R) >dp4.chr2 4266079 77 - 30794189 --CAAUUGUCAUAGUUUCUGAUGAGCUCAAUUUUGAACGCAAUGAAACCGUGACGCAUUGAGACAUCCAUCUGUCAAUA-------------------------- --(((((((((..(((((...((.(.(((....))).).))..)))))..))))).)))).((((......))))....-------------------------- ( -19.20, z-score = -1.80, R) >droPer1.super_7 4359139 77 + 4445127 --CAAUUGUCAUAGUUUCUGAUGAGCUCAAUUUUGAACGCAAUGAAACCGUGACGCAUUGAGACAUCCAUCUGUCAAUA-------------------------- --(((((((((..(((((...((.(.(((....))).).))..)))))..))))).)))).((((......))))....-------------------------- ( -19.20, z-score = -1.80, R) >droWil1.scaffold_181130 10494467 89 - 16660200 --AAAUCGUCACAGUGUUUGAUGAGCUUAAUCUUGAU-GUUAUAAAGCCGUGACGAAUUGAUGU--------AUUGAGUCAUCAGAUUGCAUUGUCCAUA----- --........((((((((((((((.((((((((..((-(((((......)))))..))..).).--------)))))))))))))...))))))).....----- ( -20.50, z-score = -1.08, R) >consensus __CAGUUGUCACAGUUUCUGAUGAGCCCAAUUGCGAUCGCAAUGAAACCGUGACGCAUUGUGGCCUGCG_CAAUUGAGACAUUAGCAAACAAUGCCGACAGAUUA ......((((((.(((((......((............))...))))).)))))).................................................. (-10.50 = -9.89 + -0.61)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:59:13 2011