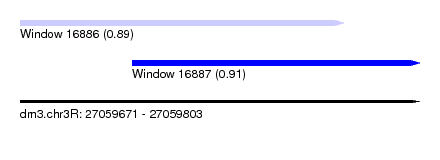
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 27,059,671 – 27,059,803 |
| Length | 132 |
| Max. P | 0.914824 |

| Location | 27,059,671 – 27,059,778 |
|---|---|
| Length | 107 |
| Sequences | 12 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 75.47 |
| Shannon entropy | 0.52974 |
| G+C content | 0.52581 |
| Mean single sequence MFE | -38.36 |
| Consensus MFE | -20.02 |
| Energy contribution | -20.19 |
| Covariance contribution | 0.18 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.888442 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
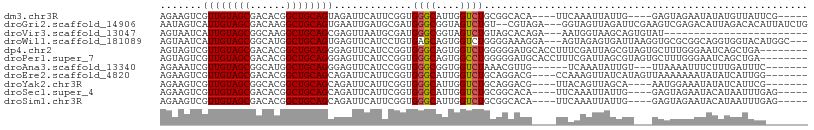
>dm3.chr3R 27059671 107 + 27905053 ---GCCAGCGGGUUGUUGUGGCACUUCUUCUGCAUCGAGAAGAAGUCGUUGUAGCGACACGGCUGCAGUAGAUUCAUUCGGUGGGCAUUGGUCUGCGGCACAUUCAAAUU---- ---((((((.((((((((..(((((((((((......))))))))).))..))))))).).)))...((((((..((((....)).))..)))))))))...........---- ( -41.80, z-score = -2.93, R) >droGri2.scaffold_14906 89383 103 - 14172833 ---GCCAGUGGAUUUUUAUGGCAAGUUUUCUGCAUUGAGAAAUAGUCAUUGUAGCGACAAGGCUGCAGUGAAUUGAUGCGAUGGGCGGUAGUCUGU-CGUAGAGGUA------- ---((((...........))))...(((((......)))))....((((((((((......))))))))))(((..((((((((((....))))))-))))..))).------- ( -35.60, z-score = -3.22, R) >droMoj3.scaffold_6540 29718882 110 - 34148556 ---GCCAGUGGAUUCUUGUGGCACUUCUUCUGCAUGGUGAAGUAGUCGUUGUAGCGACAAGGCUGCAGCGAGUUGAUGCGAUGGGCGGUCGUCUGGGCGGGCGGGCAUGCAAA- ---(((........((..(.(((((((.((.....)).)))))..((((((((((......))))))))))......)).)..))((.((((....)))).))))).......- ( -40.10, z-score = -0.30, R) >droVir3.scaffold_13047 18694895 108 - 19223366 CUCGCCAGCGGGCUUUUGUAGCAAUUUUUCUGCAUGGAGAAGUAAUCAUUGUAGCGGCAAGGCUGCAGCGAGUUAAUGCGAUGGGCGGUAGUCUGU-AGCACAGAAAUG----- (((((..((((.(((((((.(((((((((((....))))))......))))).)))).))).)))).)))))....(((.((((((....))))))-.)))((....))----- ( -33.20, z-score = -0.26, R) >droWil1.scaffold_181089 3128466 110 + 12369635 ---GCCAGUGGAUUAUUGCUGCAUUUCUUUUGCAUCGAGAAGUAAUCAUUGUAGCGGCAUGGCUGCAGUGAGUUCAUCCUGUGAGCAGUGGUCUGGGGAAAGGAAGUAGAGUG- ---.((..(((((((((((((((((((((.......)))))))..((((((((((......)))))))))).........)).)))))))))))).))...............- ( -34.30, z-score = -1.00, R) >dp4.chr2 4251561 111 + 30794189 ---GCCAGCGGAUUGUUCUGGCACUUUUUCUGCACGGAAAAGUAGUCGUUGUAGCGACACGGCUGCAGGGAGUUCAUCCGGUGGGCAGUGGUCUGGGGGAUGCACCUUUCGAUU ---.(((.(((((..(((((((((((((((.....)))))))).))).(((((((......)))))))))))...))))).))).....((((..((((.....))))..)))) ( -37.60, z-score = -0.30, R) >droPer1.super_7 4344622 111 - 4445127 ---GCCAGCGGAUUGUUCUGGCACUUUUUCUGCACGGAAAAGUAGUCGUUGUAGCGACACGGCUGCAGGGAGUUCAUCCGGUGGGCAGUGGCCUGGGGGAUGCACCUUUCGAUU ---(((((.........)))))((((((((.....))))))))((((..((((((......))))))(..((..(((((..(((((....)))))..)))))...))..))))) ( -39.40, z-score = -0.47, R) >droAna3.scaffold_13340 4941574 108 + 23697760 ---GCAAGCGGAUUGUUGUGGCAUUUGUUCUUCAUGGAGAAGAAAUCGUUGUAGCGGCAUGGCUGCAGGGAGUUCAUCCGGUGGGCGGUGGUCUAAACGUUGUCAAAUAUU--- ---...((((....((((..((((((.(((((....))))).)))).))..))))(((.((.(..(..(((.....))).)..).))...)))....))))..........--- ( -31.00, z-score = -0.72, R) >droEre2.scaffold_4820 9549078 107 - 10470090 ---GCCAGCGGGUUGUUGUGACACUUCUUCUGCAUCGAGAAGAAGUCGUUGUAGCGACACGGCUGCAGCAGAUUCAUUCGGUGGGCAUUGGUCUGCAGGACGCCAAAGUU---- ---((((((.((((((((..(((((((((((......))))))))).))..))))))).).))))..))..........((((.(((......)))....))))......---- ( -41.70, z-score = -2.11, R) >droYak2.chr3R 27996332 107 + 28832112 ---GCCAGCGGAUUGUUGUGGCACUUCUUCUGCAUGGAGAAGAAGUCGUUGUAGCGGCACGGCUGCAGCAGAUUCAUUCGGUGGGCAUUGGUCUGCAGGACGUUACAGUU---- ---.(((.((((((((((..((.(((((((.....)))))))..(((((....)))))...))..))))).....))))).)))......(((.....))).........---- ( -38.00, z-score = -1.06, R) >droSec1.super_4 5877626 107 + 6179234 ---GCCAGCGGGUUGUUGUGGCACUUCUUCUGCAUCGAGAAGAAGUCGUUGUAGCGACACGGCUGCAGCAGAUUCAUUCGGUGGGCAUUGGUCUGCGGCACAUUCAAAUU---- ---((((((.((((((((..(((((((((((......))))))))).))..))))))).).)))...((((((..((((....)).))..)))))))))...........---- ( -43.80, z-score = -3.19, R) >droSim1.chr3R 26702843 107 + 27517382 ---GCCAGCGGGUUGUUGUGGCACUUCUUCUGCAUCGAGAAGAAGUCGUUGUAGCGACACGGCUGCAGCAGAUUCAUUCGGUGGGCAUUGGUCUGCGGCACAUUCAAAUU---- ---((((((.((((((((..(((((((((((......))))))))).))..))))))).).)))...((((((..((((....)).))..)))))))))...........---- ( -43.80, z-score = -3.19, R) >consensus ___GCCAGCGGAUUGUUGUGGCACUUCUUCUGCAUGGAGAAGAAGUCGUUGUAGCGACACGGCUGCAGCGAAUUCAUUCGGUGGGCAGUGGUCUGCGGGACGUUAAAUUU____ ...((((.(((((.........(((.(((((......))))).))).((((((((......))))))))......))))).)).))............................ (-20.02 = -20.19 + 0.18)

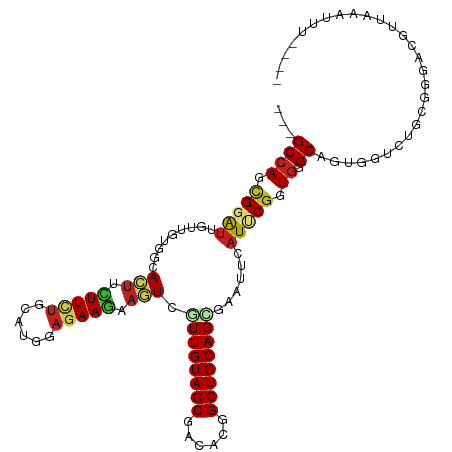
| Location | 27,059,708 – 27,059,803 |
|---|---|
| Length | 95 |
| Sequences | 11 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 61.54 |
| Shannon entropy | 0.81232 |
| G+C content | 0.48153 |
| Mean single sequence MFE | -26.66 |
| Consensus MFE | -11.82 |
| Energy contribution | -12.01 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.25 |
| Mean z-score | -0.89 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.914824 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 27059708 95 + 27905053 AGAAGUCGUUGUAGCGACACGGCUGCAGUAGAUUCAUUCGGUGGGCAUUGGUCUGCGGCACA----UUCAAAUUAUUG----GAGUAGAAUAUAUGUUAUUCG----- ....(((((....)))))...(((((((..(((((((...))))..)))...)))))))((.----(((((....)))----)))).(((((.....))))).----- ( -23.40, z-score = -0.61, R) >droGri2.scaffold_14906 89420 103 - 14172833 AAUAGUCAUUGUAGCGACAAGGCUGCAGUGAAUUGAUGCGAUGGGCGGUAGUCUGU--CGUAGA---GGUAGUUAGAUUCGAAGUCGAGACAUUAGACACAUUAUCUG .....((((((((((......))))))))))(((..((((((((((....))))))--))))..---)))...(((((.....(((.........))).....))))) ( -32.50, z-score = -2.60, R) >droVir3.scaffold_13047 18694935 81 - 19223366 AGUAAUCAUUGUAGCGGCAAGGCUGCAGCGAGUUAAUGCGAUGGGCGGUAGUCUGUAGCACAGA---AAUGGUAAGCAGUGUAU------------------------ ......((((((.((.(((.((((((.((..(((.....)))..)).))))))))).)).((..---..))....))))))...------------------------ ( -23.10, z-score = -1.03, R) >droWil1.scaffold_181089 3128503 102 + 12369635 AGUAAUCAUUGUAGCGGCAUGGCUGCAGUGAGUUCAUCCUGUGAGCAGUGGUCUGGGGAAAGGA---AGUAGAGUGAUUAAGGUGCGCGGCAGGUGGUACAUGGC--- .....((((((((((......))))))))))((((((((((((.((..(((((...........---........)))))..)).)))))..))))).)).....--- ( -28.81, z-score = -0.63, R) >dp4.chr2 4251598 100 + 30794189 AGUAGUCGUUGUAGCGACACGGCUGCAGGGAGUUCAUCCGGUGGGCAGUGGUCUGGGGGAUGCACCUUUCGAUUAGCGUAGUGCUUUGGGAAUCAGCUGA-------- ....(((((....))))).((((((..(..((..(((((..(((((....)))))..)))))...))..)....(((.....)))........)))))).-------- ( -30.40, z-score = 0.56, R) >droPer1.super_7 4344659 100 - 4445127 AGUAGUCGUUGUAGCGACACGGCUGCAGGGAGUUCAUCCGGUGGGCAGUGGCCUGGGGGAUGCACCUUUCGAUUAGCGUAGUGCUUUGGGAAUCAGCUGA-------- ....(((((....))))).((((((..(..((..(((((..(((((....)))))..)))))...))..)....(((.....)))........)))))).-------- ( -33.10, z-score = 0.16, R) >droAna3.scaffold_13340 4941611 92 + 23697760 AGAAAUCGUUGUAGCGGCAUGGCUGCAGGGAGUUCAUCCGGUGGGCGGUGGUCUAAACGUUG------UCAAAUAUUGU---UUAAAAUUUCUUUGAUUUC------- .((((((((((((((......)))))))(((.....)))....(((..((.......))..)------)).........---............)))))))------- ( -21.10, z-score = -0.55, R) >droEre2.scaffold_4820 9549115 97 - 10470090 AGAAGUCGUUGUAGCGACACGGCUGCAGCAGAUUCAUUCGGUGGGCAUUGGUCUGCAGGACG----CCAAAGUUAUCAUAGUUAAAAAAAUAUAUCAUUGG------- .((....((((((((......)))))))).(((...((.((((.(((......)))....))----)).))...))).................)).....------- ( -22.80, z-score = -0.59, R) >droYak2.chr3R 27996369 93 + 28832112 AGAAGUCGUUGUAGCGGCACGGCUGCAGCAGAUUCAUUCGGUGGGCAUUGGUCUGCAGGACG----UUACAGUUAGCA----AAUGGAAAUAUAUCAUUCG------- .((((((((((((((......))))))))...........(..(((....)))..)..)))(----(((....)))).----...............))).------- ( -24.10, z-score = -0.63, R) >droSec1.super_4 5877663 95 + 6179234 AGAAGUCGUUGUAGCGACACGGCUGCAGCAGAUUCAUUCGGUGGGCAUUGGUCUGCGGCACA----UUCAAAUUAUUG----GAGUAGAAUACAUAAUUUGAG----- ....(((((((((((......))))))))...........(..(((....)))..))))...----.(((((((((.(----..........)))))))))).----- ( -27.00, z-score = -1.90, R) >droSim1.chr3R 26702880 95 + 27517382 AGAAGUCGUUGUAGCGACACGGCUGCAGCAGAUUCAUUCGGUGGGCAUUGGUCUGCGGCACA----UUCAAAUUAUUG----GAGUAGAAUACAUAAUUUGAG----- ....(((((((((((......))))))))...........(..(((....)))..))))...----.(((((((((.(----..........)))))))))).----- ( -27.00, z-score = -1.90, R) >consensus AGAAGUCGUUGUAGCGACACGGCUGCAGCGAAUUCAUUCGGUGGGCAGUGGUCUGCGGCACG____UUAAAAUUAGCGU___GAGUAGAAUAUAUGAUUUG_______ .......((((((((......)))))))).............((((....))))...................................................... (-11.82 = -12.01 + 0.19)
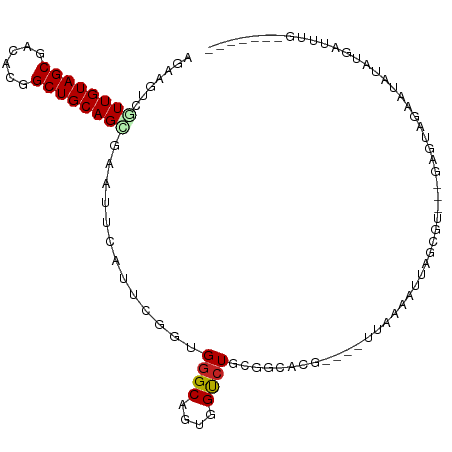
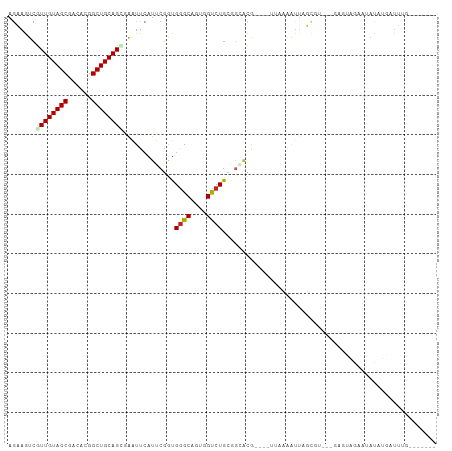
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:59:12 2011