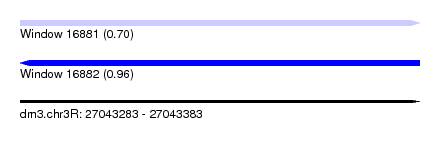
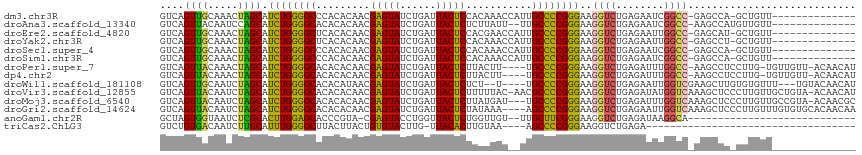
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 27,043,283 – 27,043,383 |
| Length | 100 |
| Max. P | 0.960557 |

| Location | 27,043,283 – 27,043,383 |
|---|---|
| Length | 100 |
| Sequences | 14 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 72.82 |
| Shannon entropy | 0.58230 |
| G+C content | 0.49360 |
| Mean single sequence MFE | -27.71 |
| Consensus MFE | -18.44 |
| Energy contribution | -18.29 |
| Covariance contribution | -0.15 |
| Combinations/Pair | 1.24 |
| Mean z-score | -0.46 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.702824 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
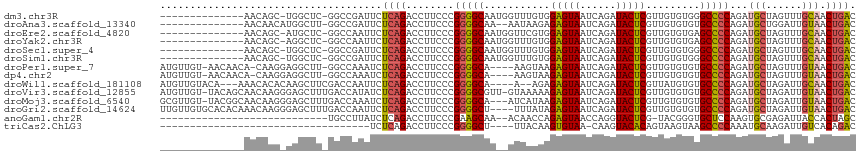
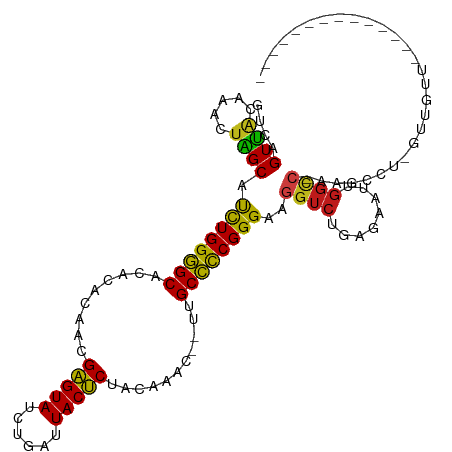
>dm3.chr3R 27043283 100 + 27905053 --------------AACAGC-UGGCUC-GGCCGAUUCUCAGACCUUCCCGGGGCAAUGGUUUGUGGAGUAAUCAGAUACUCGUUGUGUGGGCCCCAGAUGCUAGUUUGCAACUGAC --------------..((((-(((...-((((.((.(.((((((.((....))....)))))).)(((((......))))).....)).)))))))).(((......))).))).. ( -29.30, z-score = 0.56, R) >droAna3.scaffold_13340 14434699 99 + 23697760 --------------AACAACAUGGCUU-GGCCGAUUCUCAGACCUUCCCGGGGCAA--AAUAAGAGAGUAAUCAGAUACUCGUUGUGUGUGCCCCAGAUGCUGGAUUGUAACUGAC --------------.((((..((((..-.))))..((.(((....((..((((((.--.((((..(((((......))))).))))...)))))).))..)))))))))....... ( -28.10, z-score = -1.09, R) >droEre2.scaffold_4820 9532348 100 - 10470090 --------------AACAGC-AUGCUC-GGCCAAUUCUCAGACCUUCCCGGGGCAAUGGUUCGUGGAGUAAUCAGAUACUCGUUGUGUGAGCCCCAGAUGCUAGUUUGCAACUGAC --------------.(((((-...(.(-(((((..((((.(......).))))...))).))).)(((((......))))))))))........(((.(((......))).))).. ( -23.40, z-score = 1.42, R) >droYak2.chr3R 27979653 100 + 28832112 --------------AACAGC-AGGCUC-GGCCAAUUCUCAGACCUUCCCGGGGCAAUGGUUUGUGGAGUAAUCAGAUACUCGUUGUGUGAGCCCCAGAUGCUAGUUUGCAACUGAC --------------......-.(((((-(..((((.(.((((((.((....))....)))))).)(((((......)))))))))..)))))).(((.(((......))).))).. ( -30.40, z-score = -0.39, R) >droSec1.super_4 5861277 100 + 6179234 --------------AACAGC-UGGCUC-GGCCGAUUCUCAGACCUUCCCGGGGCAAUGGUUUGUGGAGUAAUCAGAUACUCGUUGUGUGGGCCCCAGAUGCUAGUUUGCAACUGAC --------------..((((-(((...-((((.((.(.((((((.((....))....)))))).)(((((......))))).....)).)))))))).(((......))).))).. ( -29.30, z-score = 0.56, R) >droSim1.chr3R 26686454 100 + 27517382 --------------AACAGC-UGGCUC-GGCCGAUUCUCAGACCUUCCCGGGGCAAUGGUUUGUGGAGUAAUCAGAUACUCGUUGUGUGGGCCCCAGAUGCUAGUUUGCAACUGAC --------------..((((-(((...-((((.((.(.((((((.((....))....)))))).)(((((......))))).....)).)))))))).(((......))).))).. ( -29.30, z-score = 0.56, R) >droPer1.super_7 1566407 109 - 4445127 AUGUUGU-AACAACA-CAAGGAGGCUU-GGCCAAAUCUCAGACCUUCCCGGGGCA----AAGUAAGAGUAAUCAGAUACUCGUUGUGUGUGCCCCAGAUGCUAGUUUGUAACUGAC .....((-.((((.(-(..(((((..(-((.......)))..)))))..((((((----......(((((......)))))........))))))........)))))).)).... ( -28.14, z-score = -0.17, R) >dp4.chr2 30707174 109 - 30794189 AUGUUGU-AACAACA-CAAGGAGGCUU-GGCCAAAUCUCAGACCUUCCCGGGGCA----AAGUAAGAGUAAUCAGAUACUCGUUGUGUGUGCCCCAGAUGCUAGUUUGUAACUGAC .....((-.((((.(-(..(((((..(-((.......)))..)))))..((((((----......(((((......)))))........))))))........)))))).)).... ( -28.14, z-score = -0.17, R) >droWil1.scaffold_181108 710415 107 + 4707319 AUGUUGUACA---AAACACACAAGCUUCGACCAAUUCUCAGACCUUCCCGGGGCA----A--AGAGAGUAAUCAGAUACUCGUUAUGUGUGCCCCAGAUGCUAGAUUGCAACUGAC ..........---...((((((.((((((...................)))))).----.--...(((((......)))))....))))))...(((.(((......))).))).. ( -25.11, z-score = -0.84, R) >droVir3.scaffold_12855 8932326 114 - 10161210 AUGUUGU-UACAGCAACAAGGGAGCUUUGACCAUAUCUCAGACCUUCCCGGGGCGUU-GUAAAAAGAGUAAUCAGAUACUCGUUGUGUGUGCCCCAGAUGCUAGAUUGUAACUGAC .....((-(((((((.(..(((((.(((((.......))))).))))).((((((..-.......(((((......)))))........)))))).).))).....)))))).... ( -32.33, z-score = -1.41, R) >droMoj3.scaffold_6540 7172155 112 - 34148556 GCGUUGU-UACGGCAACAAGGGAGCUUUGACCAAAUCUCAGACCUUCCCGGGGCA---AUCAUAAGAGUAAUCAGAUACUCGUUGUGUGUGCCCCAGAUGCUAGAUUGUAACUGAC ..(((((-(((((((.(..(((((.(((((.......))))).))))).((((((---..((((((((((......))))).)))))..)))))).).)))).....))))).))) ( -38.40, z-score = -3.01, R) >droGri2.scaffold_14624 417492 112 - 4233967 UUGUUGUGCACACAAACAAGGGAGCUUUGACCAAUUCUCAGACCUUCCCGGGGCU----UUUAUAGAGUAAUCAGAUACUCGUUGUGUGUGCCCCAGAUGCUAGAUUGUAACUGAC .....(..(((((((.(..(((((.(((((.......))))).)))))..)....----......(((((......))))).)))))))..)..(((.(((......))).))).. ( -32.00, z-score = -1.39, R) >anoGam1.chr2R 17983806 85 - 62725911 ----------------------------UGCCUUAUCUCAGACCUUCCCGAAGCAA--ACAACCAGAGUAACCAGGUACUCG-UACGGGUGCUCCAAGUGCGAGAUUACCACUAGC ----------------------------(((((...(((.................--.......))).....)))))((((-(((.((....))..)))))))............ ( -15.86, z-score = 0.69, R) >triCas2.ChLG3 160638 76 + 32080666 -----------------------------------UCUCAGACCUUCCCGGGGCU----UUACAAGUGUAA-CAAGUACACAGUAAGUAAGCCCCAAAUGCAAGAUUGUCACAGAC -----------------------------------.....(((..((..((((((----((((..(((((.-....))))).)))...)))))))........))..)))...... ( -18.10, z-score = -1.80, R) >consensus ______________AACAAC_AGGCUU_GGCCAAUUCUCAGACCUUCCCGGGGCAA__GUUAGUAGAGUAAUCAGAUACUCGUUGUGUGUGCCCCAGAUGCUAGAUUGCAACUGAC .....................................((((........(((((...........(((((......))))).........)))))...(((......))).)))). (-18.44 = -18.29 + -0.15)
| Location | 27,043,283 – 27,043,383 |
|---|---|
| Length | 100 |
| Sequences | 14 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 72.82 |
| Shannon entropy | 0.58230 |
| G+C content | 0.49360 |
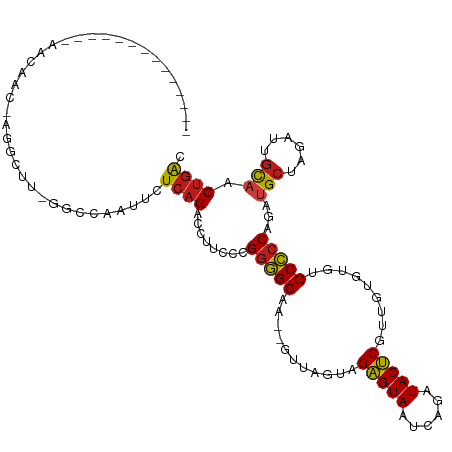
| Mean single sequence MFE | -29.75 |
| Consensus MFE | -20.81 |
| Energy contribution | -20.07 |
| Covariance contribution | -0.74 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.05 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.68 |
| SVM RNA-class probability | 0.960557 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 27043283 100 - 27905053 GUCAGUUGCAAACUAGCAUCUGGGGCCCACACAACGAGUAUCUGAUUACUCCACAAACCAUUGCCCCGGGAAGGUCUGAGAAUCGGCC-GAGCCA-GCUGUU-------------- ..(((((((.........((((((((.........(((((......)))))...........))))))))..((((........))))-..).))-))))..-------------- ( -30.35, z-score = -0.86, R) >droAna3.scaffold_13340 14434699 99 - 23697760 GUCAGUUACAAUCCAGCAUCUGGGGCACACACAACGAGUAUCUGAUUACUCUCUUAUU--UUGCCCCGGGAAGGUCUGAGAAUCGGCC-AAGCCAUGUUGUU-------------- .......(((((...((.(((((((((........(((((......))))).......--.)))))))))..((((........))))-..))...))))).-------------- ( -30.39, z-score = -1.86, R) >droEre2.scaffold_4820 9532348 100 + 10470090 GUCAGUUGCAAACUAGCAUCUGGGGCUCACACAACGAGUAUCUGAUUACUCCACGAACCAUUGCCCCGGGAAGGUCUGAGAAUUGGCC-GAGCAU-GCUGUU-------------- ..(((((((.........((((((((.........(((((......)))))...........))))))))..((((........))))-..))).-))))..-------------- ( -31.65, z-score = -1.16, R) >droYak2.chr3R 27979653 100 - 28832112 GUCAGUUGCAAACUAGCAUCUGGGGCUCACACAACGAGUAUCUGAUUACUCCACAAACCAUUGCCCCGGGAAGGUCUGAGAAUUGGCC-GAGCCU-GCUGUU-------------- ..((((.((.........((((((((.........(((((......)))))...........))))))))..((((........))))-..))..-))))..-------------- ( -30.65, z-score = -0.91, R) >droSec1.super_4 5861277 100 - 6179234 GUCAGUUGCAAACUAGCAUCUGGGGCCCACACAACGAGUAUCUGAUUACUCCACAAACCAUUGCCCCGGGAAGGUCUGAGAAUCGGCC-GAGCCA-GCUGUU-------------- ..(((((((.........((((((((.........(((((......)))))...........))))))))..((((........))))-..).))-))))..-------------- ( -30.35, z-score = -0.86, R) >droSim1.chr3R 26686454 100 - 27517382 GUCAGUUGCAAACUAGCAUCUGGGGCCCACACAACGAGUAUCUGAUUACUCCACAAACCAUUGCCCCGGGAAGGUCUGAGAAUCGGCC-GAGCCA-GCUGUU-------------- ..(((((((.........((((((((.........(((((......)))))...........))))))))..((((........))))-..).))-))))..-------------- ( -30.35, z-score = -0.86, R) >droPer1.super_7 1566407 109 + 4445127 GUCAGUUACAAACUAGCAUCUGGGGCACACACAACGAGUAUCUGAUUACUCUUACUU----UGCCCCGGGAAGGUCUGAGAUUUGGCC-AAGCCUCCUUG-UGUUGUU-ACAACAU ....((.((((....((((((((((((........(((((......)))))......----)))))))))..((((........))))-.........))-).)))).-))..... ( -31.04, z-score = -1.28, R) >dp4.chr2 30707174 109 + 30794189 GUCAGUUACAAACUAGCAUCUGGGGCACACACAACGAGUAUCUGAUUACUCUUACUU----UGCCCCGGGAAGGUCUGAGAUUUGGCC-AAGCCUCCUUG-UGUUGUU-ACAACAU ....((.((((....((((((((((((........(((((......)))))......----)))))))))..((((........))))-.........))-).)))).-))..... ( -31.04, z-score = -1.28, R) >droWil1.scaffold_181108 710415 107 - 4707319 GUCAGUUGCAAUCUAGCAUCUGGGGCACACAUAACGAGUAUCUGAUUACUCUCU--U----UGCCCCGGGAAGGUCUGAGAAUUGGUCGAAGCUUGUGUGUUU---UGUACAACAU .((((.(((......)))(((((((((........(((((......)))))...--.----))))))))).....))))...(((..((((((......))))---))..)))... ( -30.70, z-score = -0.95, R) >droVir3.scaffold_12855 8932326 114 + 10161210 GUCAGUUACAAUCUAGCAUCUGGGGCACACACAACGAGUAUCUGAUUACUCUUUUUAC-AACGCCCCGGGAAGGUCUGAGAUAUGGUCAAAGCUCCCUUGUUGCUGUA-ACAACAU ..((((.((((...(((.((((((((.........(((((......))))).......-...))))))))..(..(........)..)...)))...)))).))))..-....... ( -31.61, z-score = -1.65, R) >droMoj3.scaffold_6540 7172155 112 + 34148556 GUCAGUUACAAUCUAGCAUCUGGGGCACACACAACGAGUAUCUGAUUACUCUUAUGAU---UGCCCCGGGAAGGUCUGAGAUUUGGUCAAAGCUCCCUUGUUGCCGUA-ACAACGC ....((((((((((.((.(((((((((..((.((.(((((......))))))).))..---)))))))))...))...))))).(((..(((....)))...))))))-))..... ( -33.80, z-score = -1.74, R) >droGri2.scaffold_14624 417492 112 + 4233967 GUCAGUUACAAUCUAGCAUCUGGGGCACACACAACGAGUAUCUGAUUACUCUAUAAA----AGCCCCGGGAAGGUCUGAGAAUUGGUCAAAGCUCCCUUGUUUGUGUGCACAACAA ....(((....(((((...)))))(((((((.((((((((......)))))......----......((((.(.(.(((.......))).).)))))..))))))))))..))).. ( -27.70, z-score = -0.12, R) >anoGam1.chr2R 17983806 85 + 62725911 GCUAGUGGUAAUCUCGCACUUGGAGCACCCGUA-CGAGUACCUGGUUACUCUGGUUGU--UUGCUUCGGGAAGGUCUGAGAUAAGGCA---------------------------- ....((((.....)))).(((((((((..((..-((((((......))))).)..)).--.)))))))))...((((......)))).---------------------------- ( -23.20, z-score = 0.61, R) >triCas2.ChLG3 160638 76 - 32080666 GUCUGUGACAAUCUUGCAUUUGGGGCUUACUUACUGUGUACUUG-UUACACUUGUAA----AGCCCCGGGAAGGUCUGAGA----------------------------------- .(((..(((..(((......((((((((...(((.(((((....-.)))))..))))----))))))))))..)))..)))----------------------------------- ( -23.60, z-score = -1.83, R) >consensus GUCAGUUACAAACUAGCAUCUGGGGCACACACAACGAGUAUCUGAUUACUCUACAAAC__UUGCCCCGGGAAGGUCUGAGAAUUGGCC_AAGCCU_GUUGUU______________ ....((((.....)))).((((((((.........(((((......)))))...........))))))))..((((........))))............................ (-20.81 = -20.07 + -0.74)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:59:08 2011