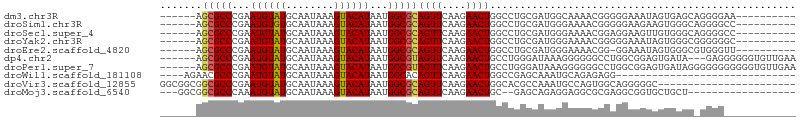
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 27,013,787 – 27,013,877 |
| Length | 90 |
| Max. P | 0.995359 |

| Location | 27,013,787 – 27,013,877 |
|---|---|
| Length | 90 |
| Sequences | 10 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 67.83 |
| Shannon entropy | 0.63530 |
| G+C content | 0.52973 |
| Mean single sequence MFE | -21.86 |
| Consensus MFE | -12.31 |
| Energy contribution | -12.45 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.79 |
| SVM RNA-class probability | 0.995359 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
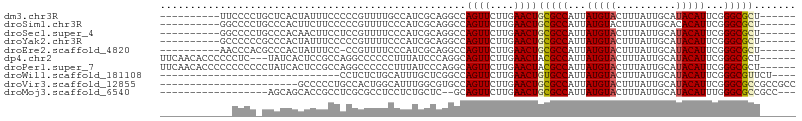
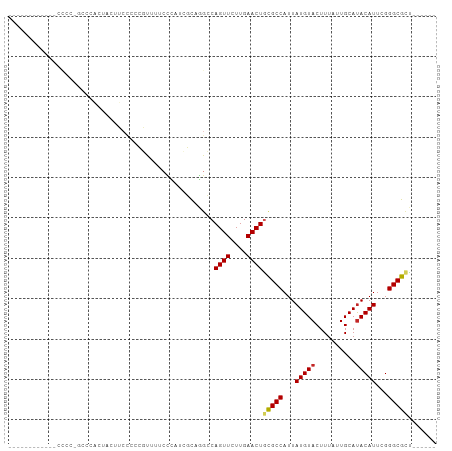
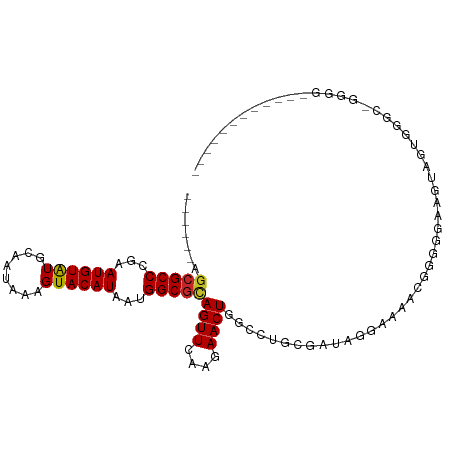
>dm3.chr3R 27013787 90 + 27905053 ----------UUCCCCUGCUCACUAUUUCCCCCGUUUUGCCAUCGCAGGCCAGUUCUUGAACUGCGCCAUUAUGUACUUUAUUGCAUACAUUCGGGCGCU------ ----------.......((((................(((....)))((((((((....))))).)))..((((((......)))))).....))))...------ ( -21.00, z-score = -1.60, R) >droSim1.chr3R 26654922 90 + 27517382 ----------GGCCCCUGCCCACUUCUUCCCCCGUUUUCCCAUCGCAGGCCAGUUCUUGAACUGCGCCAUUAUGUACUUUAUUGCACACAUUCGGGCGCU------ ----------.......((((..........................((((((((....))))).)))....((((......)))).......))))...------ ( -20.80, z-score = -1.01, R) >droSec1.super_4 5832011 90 + 6179234 ----------GGCCCCUGCCCACAACUUCCUCCGUUUUCCCAUCGCAGGCCAGUUCUUGAACUGCGCCAUUAUGUACUUUAUUGCAUACAUUCGGGCGCU------ ----------.......((((..........................((((((((....))))).)))..((((((......)))))).....))))...------ ( -23.10, z-score = -1.81, R) >droYak2.chr3R 27949746 90 + 28832112 ----------GCCCCCCGCCCACUAUUUCCCCCGUUUUCCCAUCGCAGGCCAGUUCUUGAACUGCGCCAUUAUGUACUUUAUUGCAUACAUUCGGGCGCU------ ----------......(((((..........................((((((((....))))).)))..((((((......)))))).....)))))..------ ( -24.50, z-score = -3.44, R) >droEre2.scaffold_4820 9501622 89 - 10470090 ----------AACCCACGCCCACUAUUUCC-CCGUUUUCCCAUCGCAGGCCAGUUCUUGAACUGCGCCAUUAUGUACUUUAUUGCAUACAUUCGGGCGCU------ ----------......(((((.........-................((((((((....))))).)))..((((((......)))))).....)))))..------ ( -24.70, z-score = -3.70, R) >dp4.chr2 30676035 97 - 30794189 UUCAACACCCCCCUC---UAUCACUCCGCCAGGCCCCCCUUUAUCCCAGGCAGUUCUUGAACUACGCCAUUAUGUACUUUAUUGCAUACAUUCGGGCGCU------ ...............---..............((((............(((((((....))))..)))..((((((......)))))).....))))...------ ( -17.30, z-score = -0.87, R) >droPer1.super_7 1535281 100 - 4445127 UUCAACACCCCCCCCCCCUAUCACUCCGCCAGGCCCCCCUUUAUCCCAGGCAGUUCUUGAACUACGCCAUUAUGUACUUUAUUGCAUACAUUCGGGCGCU------ ................................((((............(((((((....))))..)))..((((((......)))))).....))))...------ ( -17.30, z-score = -0.96, R) >droWil1.scaffold_181108 680219 72 + 4707319 ------------------------------CCUCUCUGCAUUUGCUCGGCCAGUUCUUGAACUGUGCCAUUAUGUACUUUAUUGCAUACAUUCGGGCGUUCU---- ------------------------------.............((((((((((((....))))).)))..((((((......)))))).....)))).....---- ( -20.20, z-score = -3.00, R) >droVir3.scaffold_12855 8896617 83 - 10161210 -----------------------GCCCCCUGCCACUGGCAUUUGGCGUGCCAGUUCUUGAACUGCGCCAUUAUGUACUUUAUUGCAUACAUUCGGGCGCCGCCGCC -----------------------((....((((...))))...((((.(((((((....))))).(((...(((((..........)))))...))))))))))). ( -25.90, z-score = -0.83, R) >droMoj3.scaffold_6540 7136169 83 - 34148556 ------------------AGCAGCACCGCCUCGCGCCUCCUCUGCUC--GCAGUUCUUGAACUGCGCCAUUAUGUACUUUAUUGCAUACAUUUGGGCGCCGCC--- ------------------.........((...((((((........(--((((((....)))))))....((((((......)))))).....)))))).)).--- ( -23.80, z-score = -1.05, R) >consensus ____________CCCC_GCCCACUACUUCCCCCGUUUUCCCAUCGCAGGCCAGUUCUUGAACUGCGCCAUUAUGUACUUUAUUGCAUACAUUCGGGCGCU______ ...................................................((((....))))(((((...(((((..........)))))...)))))....... (-12.31 = -12.45 + 0.14)
| Location | 27,013,787 – 27,013,877 |
|---|---|
| Length | 90 |
| Sequences | 10 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 67.83 |
| Shannon entropy | 0.63530 |
| G+C content | 0.52973 |
| Mean single sequence MFE | -26.86 |
| Consensus MFE | -14.69 |
| Energy contribution | -14.66 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.955355 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
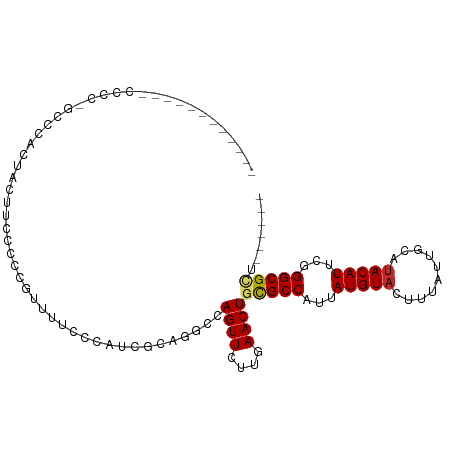
>dm3.chr3R 27013787 90 - 27905053 ------AGCGCCCGAAUGUAUGCAAUAAAGUACAUAAUGGCGCAGUUCAAGAACUGGCCUGCGAUGGCAAAACGGGGGAAAUAGUGAGCAGGGGAA---------- ------.(((((...((((((........))))))...))))).((((....((((.(((.((.........)).)))...)))))))).......---------- ( -25.90, z-score = -1.26, R) >droSim1.chr3R 26654922 90 - 27517382 ------AGCGCCCGAAUGUGUGCAAUAAAGUACAUAAUGGCGCAGUUCAAGAACUGGCCUGCGAUGGGAAAACGGGGGAAGAAGUGGGCAGGGGCC---------- ------...((((...(((((((......)))))))...((.((.(((.....(((.(((.....)))....))).....))).)).))..)))).---------- ( -29.20, z-score = -1.56, R) >droSec1.super_4 5832011 90 - 6179234 ------AGCGCCCGAAUGUAUGCAAUAAAGUACAUAAUGGCGCAGUUCAAGAACUGGCCUGCGAUGGGAAAACGGAGGAAGUUGUGGGCAGGGGCC---------- ------.(((((...((((((........))))))...))))).(((((..((((..(((.((.........)).))).)))).))))).......---------- ( -28.70, z-score = -1.50, R) >droYak2.chr3R 27949746 90 - 28832112 ------AGCGCCCGAAUGUAUGCAAUAAAGUACAUAAUGGCGCAGUUCAAGAACUGGCCUGCGAUGGGAAAACGGGGGAAAUAGUGGGCGGGGGGC---------- ------.(((((...((((((........))))))...))))).((((..(.((((.(((.((.........)).)))...))))...)...))))---------- ( -26.00, z-score = -1.10, R) >droEre2.scaffold_4820 9501622 89 + 10470090 ------AGCGCCCGAAUGUAUGCAAUAAAGUACAUAAUGGCGCAGUUCAAGAACUGGCCUGCGAUGGGAAAACGG-GGAAAUAGUGGGCGUGGGUU---------- ------.(((((((.((((((........))))))...(((.(((((....))))))))................-........))))))).....---------- ( -26.80, z-score = -1.31, R) >dp4.chr2 30676035 97 + 30794189 ------AGCGCCCGAAUGUAUGCAAUAAAGUACAUAAUGGCGUAGUUCAAGAACUGCCUGGGAUAAAGGGGGGCCUGGCGGAGUGAUA---GAGGGGGGUGUUGAA ------(((((((..((((((........)))))).((.((...((...((..((.(((.......))).))..)).))...)).)).---.....)))))))... ( -25.80, z-score = -0.26, R) >droPer1.super_7 1535281 100 + 4445127 ------AGCGCCCGAAUGUAUGCAAUAAAGUACAUAAUGGCGUAGUUCAAGAACUGCCUGGGAUAAAGGGGGGCCUGGCGGAGUGAUAGGGGGGGGGGGUGUUGAA ------(((((((..((((((........)))))).((.((...((...((..((.(((.......))).))..)).))...)).)).........)))))))... ( -25.80, z-score = 0.14, R) >droWil1.scaffold_181108 680219 72 - 4707319 ----AGAACGCCCGAAUGUAUGCAAUAAAGUACAUAAUGGCACAGUUCAAGAACUGGCCGAGCAAAUGCAGAGAGG------------------------------ ----.....((.(..((((((........))))))...(((.(((((....))))))))).)).............------------------------------ ( -17.10, z-score = -1.88, R) >droVir3.scaffold_12855 8896617 83 + 10161210 GGCGGCGGCGCCCGAAUGUAUGCAAUAAAGUACAUAAUGGCGCAGUUCAAGAACUGGCACGCCAAAUGCCAGUGGCAGGGGGC----------------------- ((((((.(((((...((((((........))))))...)))))((((....)))).)).))))...((((...))))......----------------------- ( -30.60, z-score = -1.19, R) >droMoj3.scaffold_6540 7136169 83 + 34148556 ---GGCGGCGCCCAAAUGUAUGCAAUAAAGUACAUAAUGGCGCAGUUCAAGAACUGC--GAGCAGAGGAGGCGCGAGGCGGUGCUGCU------------------ ---(((((((((...((((((........))))))..((.(((((((....))))))--)..)).....(.(....).))))))))))------------------ ( -32.70, z-score = -2.13, R) >consensus ______AGCGCCCGAAUGUAUGCAAUAAAGUACAUAAUGGCGCAGUUCAAGAACUGGCCUGCGAUAGGAAAACGGGGGAAGUAGUGGGC_GGGG____________ .......(((((...((((((........))))))...)))))((((....))))................................................... (-14.69 = -14.66 + -0.03)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:59:03 2011