
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 26,988,210 – 26,988,327 |
| Length | 117 |
| Max. P | 0.585771 |

| Location | 26,988,210 – 26,988,327 |
|---|---|
| Length | 117 |
| Sequences | 7 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 65.07 |
| Shannon entropy | 0.65717 |
| G+C content | 0.41633 |
| Mean single sequence MFE | -30.24 |
| Consensus MFE | -10.30 |
| Energy contribution | -10.43 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.585771 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 26988210 117 + 27905053 UGCGAAUUCUUUCCACAUUAUGCCCCUUUUGAGGAGUGGAAUUCACUUUUAAUUUCUUAGGAUAAAAAAAGGGGCAAAUAAUG---UUGCAAAUCGGCAGAUAGGGAGGGGGUGCUCAAG .(((...(((((((((((((((((((((((.(((((((.....)))))))....((....))....))))))))))..)))))---)(((......)))....)))))))..)))..... ( -36.70, z-score = -2.64, R) >droSim1.chr3R 26628943 117 + 27517382 UGCGAAUUCUUUCCGCAUUACGCCCCUUUUGAGGAGUGGAAUUCACUUUUAAUUUCGUAGGGCAACAAAGGGGGCACAUAAUG---UUGCAAAUCGGCGGAUGGGGAGGGGGUGCUCAAA .(((..((((..((((((((.(((((((((((((((((.....)))))))......(.....)..))))))))))...)))))---)(((......)))...))..))))..)))..... ( -39.90, z-score = -2.35, R) >droSec1.super_4 5806457 117 + 6179234 UGCGAAUUCUUUCCCAAUUACGCCCCUUUUGAGGAGUGGGAUUCACUUUUAAUUUCGUAGGAUAACAAAGGGGGCACAUAAUG---UUGCAAAUCGGCGGAUGGGGAGGGUGUGCUCAAG .(((...(((((((((.....(((((((((((((((((.....)))))))....((....))...))))))))))........---((((......)))).)))))))))..)))..... ( -40.20, z-score = -2.64, R) >droYak2.chr3R 27922304 117 + 28832112 UGCGAAUUCUUUCCGCAUUACGCUUCUUUUGAGAAGGGGAAUUCACUUUUAAUUUCGUAGUAUAACAAAGAAGGCACAUAAUG---UUGCAAAUCGGCGGAUGGGGAAGGGGUGCUCAAG .((..(((((((((.((((.(((((((((...((((..(((......)))..))))((......)))))))))(((.......---.)))....))...)))).)))))))))))..... ( -27.70, z-score = -0.61, R) >droEre2.scaffold_4820 9472841 104 - 10470090 UGCGAAUUCUUUCCGCAUUACGCCCCUUUUGAGAAGGGGAAUUCACUUUUAAUUUCGUAGUAUAACAAAGGGGGCAAAUAAUG---UUGCAAGUCGAUUUGCUCAAG------------- .((((((((((.(.((((((.((((((((((.((((..(((......)))..)))).........))))))))))...)))))---).).)))..))))))).....------------- ( -29.40, z-score = -2.35, R) >dp4.chr2 30648814 109 - 30794189 -----------CAUCCAUAGCUCCCCUUUUGUAGAGGAAAAUUGGUUUUUAAUUUCGAAAUAUUCCAAGGGAAAAAUGCAAGAGCUCCGGACAGUAACAAAUUGUAAAAGAAUGUCCAAA -----------.......(((((.((((.....))))....(((((((((..(((.((.....)).)))..)))))).))))))))..(((((...................)))))... ( -18.51, z-score = 0.31, R) >droPer1.super_7 1508096 109 - 4445127 -----------CGUCCAUAGCUCCACUUUUGUAGAGGAAAAUUGGUUUUUAAUUUCGAAAUAUUCCAAGGGAAAAAUGCAAGAGCUCCGGACAGUAACAAAUUGUAAAAGAAUGUCCAAA -----------.((((..(((((.....(((((........((((.((((......))))....))))........))))))))))..))))............................ ( -19.29, z-score = 0.02, R) >consensus UGCGAAUUCUUUCCCCAUUACGCCCCUUUUGAGGAGGGGAAUUCACUUUUAAUUUCGUAGUAUAACAAAGGGGGCACAUAAUG___UUGCAAAUCGGCAGAUGGGGAAGGGGUGCUCAAA .....................((((((((((......((((((.......)))))).........))))))))))............................................. (-10.30 = -10.43 + 0.13)
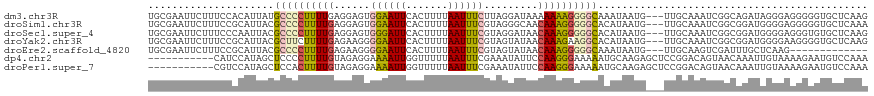

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:59:01 2011