
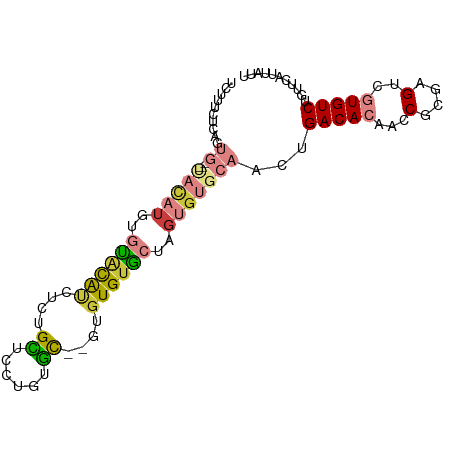
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 26,987,244 – 26,987,335 |
| Length | 91 |
| Max. P | 0.626264 |

| Location | 26,987,244 – 26,987,335 |
|---|---|
| Length | 91 |
| Sequences | 7 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 74.88 |
| Shannon entropy | 0.48504 |
| G+C content | 0.46635 |
| Mean single sequence MFE | -26.74 |
| Consensus MFE | -12.87 |
| Energy contribution | -12.86 |
| Covariance contribution | -0.01 |
| Combinations/Pair | 1.45 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.626264 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
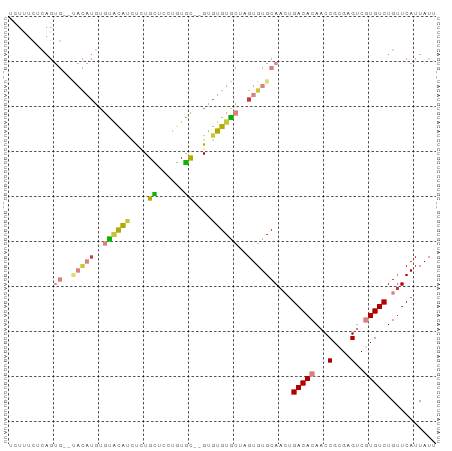
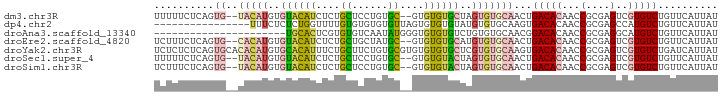
>dm3.chr3R 26987244 91 - 27905053 UUUUUCUCAGUG--UACAUGUGUACAUCUCUGCUCCUGUGC--GUGUGUGCUAGUGUGCAACUGACACAACCGCGAGUCGUGUCUGUUCAUUAUU ...........(--((((..((((((..........)))))--)..)))))(((((.(((...(((((...(....)..)))))))).))))).. ( -25.20, z-score = -1.12, R) >dp4.chr2 30647730 79 + 30794189 ----------------UUUCUCUCUGGUUUUGUGUGUGUGUUAGUGUGUGUAUGUGUGCAAGUGACACAACCGCGAGCCAUGUCUGUUCAUUAUU ----------------........((((((.(((..(((((((.((..(......)..))..)))))))..)))))))))............... ( -20.30, z-score = -1.39, R) >droAna3.scaffold_13340 14370325 73 - 23697760 ----------------------UGCACUCGUGUGUCAAUAUGGGUGUGUGUCUGUGUGCAACGGACACAACCGCGAGGCAUGUCUGUUCAUUAUU ----------------------.((((((((((....))))))))))((((((((.....))))))))..((....))................. ( -27.20, z-score = -1.94, R) >droEre2.scaffold_4820 9471769 91 + 10470090 UCUUUCUCAGUG--CACAUGUGUACAUCUCUGCUGCUAUGC--GUGUGUGCAUGUGUGCAACUGACACAACCGCGAGUCGUGUCUGUUCAUUAUU ......((((((--(((((((((((((...(((......))--).)))))))))))))).))))).(((..((((...))))..)))........ ( -32.20, z-score = -2.44, R) >droYak2.chr3R 27921349 95 - 28832112 UCUCUCUCAGUGCACACAUGUGCACAUUUCUGCUUCUGUGCGUGUGUGUGCUCGUGUGCAAGUGACACAACCGCGAGUCGUGUCUGAUCAUUAUU ......(((.(((((((..(..(((((...(((......))).)))))..)..)))))))...(((((...(....)..))))))))........ ( -31.90, z-score = -1.31, R) >droSec1.super_4 5805479 91 - 6179234 UUUUUCUCAGUG--UACAUGUGUACAUCUCUGCUCCUGUGC--GUGUGUACUAGUGUGCAACUGACACAACCGCGAGUCGUGUCUGUUCAUUAUU ...........(--((((..((((((..........)))))--)..)))))(((((.(((...(((((...(....)..)))))))).))))).. ( -25.20, z-score = -1.41, R) >droSim1.chr3R 26627979 91 - 27517382 UCUUUCUCAGUG--UACAUGUGUACAUCUCUGCUCCUGUGC--GUGUGUACUAGUGUGCAACUGACACAACCGCGAGUCGUGUCUGUUCAUUAUU ...........(--((((..((((((..........)))))--)..)))))(((((.(((...(((((...(....)..)))))))).))))).. ( -25.20, z-score = -1.33, R) >consensus UCUUUCUCAGUG__UACAUGUGUACAUCUCUGCUCCUGUGC__GUGUGUGCUAGUGUGCAACUGACACAACCGCGAGUCGUGUCUGUUCAUUAUU .....................((((((....((......))....))))))....(((.(((.(((((...(....)..))))).)))))).... (-12.87 = -12.86 + -0.01)

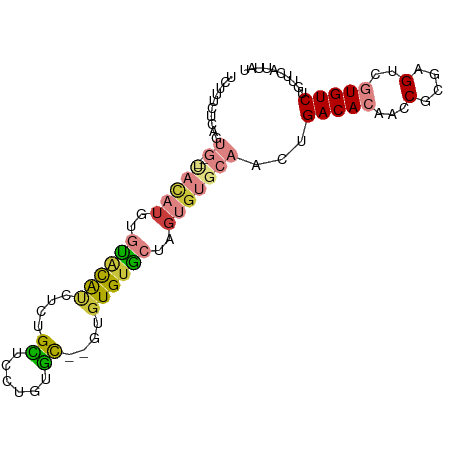
| Location | 26,987,245 – 26,987,335 |
|---|---|
| Length | 90 |
| Sequences | 7 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 74.61 |
| Shannon entropy | 0.49020 |
| G+C content | 0.47179 |
| Mean single sequence MFE | -26.74 |
| Consensus MFE | -12.87 |
| Energy contribution | -12.86 |
| Covariance contribution | -0.01 |
| Combinations/Pair | 1.45 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.585326 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 26987245 90 - 27905053 UUUUUCUCAGUG--UACAUGUGUACAUCUCUGCUCCUGUGC--GUGUGUGCUAGUGUGCAACUGACACAACCGCGAGUCGUGUCUGUUCAUUAU ...........(--((((..((((((..........)))))--)..)))))(((((.(((...(((((...(....)..)))))))).))))). ( -25.20, z-score = -1.05, R) >dp4.chr2 30647731 78 + 30794189 ----------------UUUCUCUCUGGUUUUGUGUGUGUGUUAGUGUGUGUAUGUGUGCAAGUGACACAACCGCGAGCCAUGUCUGUUCAUUAU ----------------........((((((.(((..(((((((.((..(......)..))..)))))))..))))))))).............. ( -20.30, z-score = -1.32, R) >droAna3.scaffold_13340 14370326 72 - 23697760 ----------------------UGCACUCGUGUGUCAAUAUGGGUGUGUGUCUGUGUGCAACGGACACAACCGCGAGGCAUGUCUGUUCAUUAU ----------------------.((((((((((....))))))))))((((((((.....))))))))..((....))................ ( -27.20, z-score = -1.91, R) >droEre2.scaffold_4820 9471770 90 + 10470090 UCUUUCUCAGUG--CACAUGUGUACAUCUCUGCUGCUAUGC--GUGUGUGCAUGUGUGCAACUGACACAACCGCGAGUCGUGUCUGUUCAUUAU ......((((((--(((((((((((((...(((......))--).)))))))))))))).))))).(((..((((...))))..)))....... ( -32.20, z-score = -2.33, R) >droYak2.chr3R 27921350 94 - 28832112 UCUCUCUCAGUGCACACAUGUGCACAUUUCUGCUUCUGUGCGUGUGUGUGCUCGUGUGCAAGUGACACAACCGCGAGUCGUGUCUGAUCAUUAU ......(((.(((((((..(..(((((...(((......))).)))))..)..)))))))...(((((...(....)..))))))))....... ( -31.90, z-score = -1.27, R) >droSec1.super_4 5805480 90 - 6179234 UUUUUCUCAGUG--UACAUGUGUACAUCUCUGCUCCUGUGC--GUGUGUACUAGUGUGCAACUGACACAACCGCGAGUCGUGUCUGUUCAUUAU ...........(--((((..((((((..........)))))--)..)))))(((((.(((...(((((...(....)..)))))))).))))). ( -25.20, z-score = -1.31, R) >droSim1.chr3R 26627980 90 - 27517382 UCUUUCUCAGUG--UACAUGUGUACAUCUCUGCUCCUGUGC--GUGUGUACUAGUGUGCAACUGACACAACCGCGAGUCGUGUCUGUUCAUUAU ...........(--((((..((((((..........)))))--)..)))))(((((.(((...(((((...(....)..)))))))).))))). ( -25.20, z-score = -1.21, R) >consensus UCUUUCUCAGUG__UACAUGUGUACAUCUCUGCUCCUGUGC__GUGUGUGCUAGUGUGCAACUGACACAACCGCGAGUCGUGUCUGUUCAUUAU .....................((((((....((......))....))))))....(((.(((.(((((...(....)..))))).))))))... (-12.87 = -12.86 + -0.01)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:59:00 2011