| Sequence ID | dm3.chr3R |
|---|---|
| Location | 26,984,207 – 26,984,317 |
| Length | 110 |
| Max. P | 0.923218 |

| Location | 26,984,207 – 26,984,317 |
|---|---|
| Length | 110 |
| Sequences | 11 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 74.75 |
| Shannon entropy | 0.52003 |
| G+C content | 0.64533 |
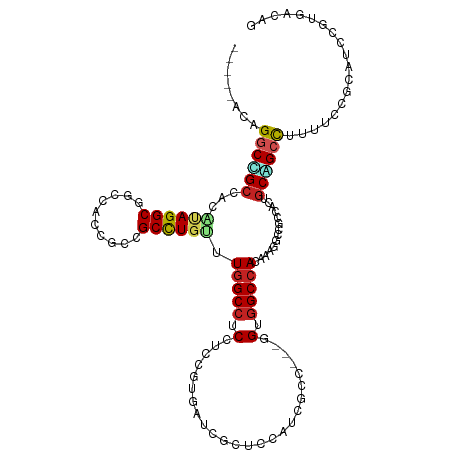
| Mean single sequence MFE | -47.75 |
| Consensus MFE | -20.39 |
| Energy contribution | -20.76 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.44 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.911071 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
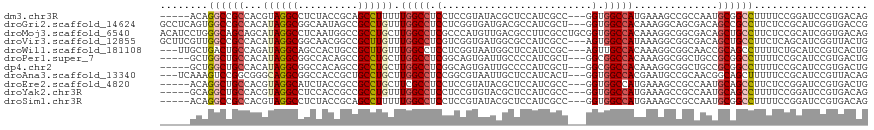
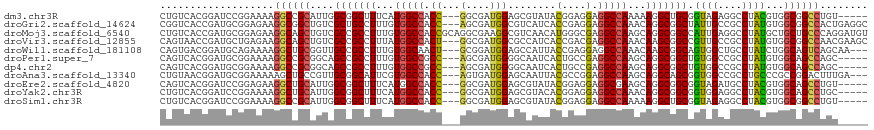
>dm3.chr3R 26984207 110 + 27905053 -----ACAGGCCGCCACGUAGGCCUCUACCGCAGCCUUUUUGGCCUCCUCCGUAUACGCUCCAUCGCC---GGUGGCCAUGAAAGCCGCCAAUGCGGCCUUUUCCGGAUCCGUGACAG -----..(((((((...((((....)))).((.((.(((.(((((.((..((............))..---)).))))).))).)).))....)))))))..((((....)).))... ( -36.70, z-score = -0.84, R) >droGri2.scaffold_14624 334503 115 - 4233967 GCCUCAGUGGCCGCCACAUAGGCGGCAAUAGCCGCCUGUUUGGCCUCCUCGGUGAUGACGCCAUCGCU---GGUGGCCACAAAGGCAGCGACAGCCGCCUUCUCCGCAUCGGUGACCG ((((..((((((((((.(((((((((....))))))))).))))....(((((((((....)))))))---)).))))))..)))).(((.....)))..((.(((...))).))... ( -60.90, z-score = -4.65, R) >droMoj3.scaffold_6540 7092033 118 - 34148556 ACAUCCUGGGCAGCAGCAUAGGCCUCAAUGGCCGCCUGCUUGGCCUCGCCCAUGUUGACGCCUUCGCCUGCGGUGGCCACAAAGGCGGCGACAGCUGCCUUCUCCGCAUCGGUGACAG .....((((((..((((((.((((.....))))(((.....))).......))))))..))).((((((((((.((......(((((((....))))))))).)))))..)))))))) ( -52.30, z-score = -1.49, R) >droVir3.scaffold_12855 8842311 115 - 10161210 GCUUCGUUGGCCGCCACAUAGGCGGCAACGGCCGCUUGUUUGGCCUCGUCGGUGAUGGCGCCAUCGCC---AGUGGCCAUAAAGGCGGCGACAGCUGCCUUCUCAGCAUCGGUUACUG (((....(((((((((.(((((((((....))))))))).))))......(((((((....)))))))---...)))))..((((((((....))))))))...)))........... ( -58.80, z-score = -3.80, R) >droWil1.scaffold_181108 644503 112 + 4707319 ---UUGCUGACUGCCAGAUAGGCAGCCACUGCCGCUUGUUUGGCCUCCUCGGUAAUGGCUCCAUCCGC---AGUUGCCACAAAGGCGGCAACCGCAGCCUUUUCUGCAUCCGUCACUG ---....(((((((.(((.((((.((...((((((((...((((..(..(((..(((....)))))).---.)..))))...))))))))...)).))))..))))))...))))... ( -42.50, z-score = -2.11, R) >droPer1.super_7 1502630 110 - 4445127 -----GCUGGCUGCCACAUAGGCGGCCACAGCCGCCUGCUUGGCCUCGGCAGUGAUUGCCCCAUCGCU---GGCGGCCACAAAGGCGGCUGCCGCGGCCUUUUCCGCAUCCGUGACUG -----..((((((((.....))))))))(((((((((...(((((..(.(((((((......))))))---).))))))...)))))))))..((((......))))........... ( -59.60, z-score = -3.26, R) >dp4.chr2 30644536 110 - 30794189 -----GCUGGCUGCCACAUAGGCGGCCACAGCCGCCUGCUUGGCCUCGGCAGUGAUUGCCCCAUCGCU---GGCGGCCACAAAGGCGGCUGCCGCGGCCUUUUCCGCAUCCGUGACUG -----..((((((((.....))))))))(((((((((...(((((..(.(((((((......))))))---).))))))...)))))))))..((((......))))........... ( -59.60, z-score = -3.26, R) >droAna3.scaffold_13340 14366740 112 + 23697760 ---UCAAAGUCCGGCGGGCAGGCGGCCACCGCUGCCUGCUUGGCCUCCGGCGUAAUUGCUCCAUCACU---GGUGGCCACGAAUGCCGCAACGGCAGCUUUUUCCGCAUCCGUUACAG ---........(((((((((((((((....)))))))))))((((.((((.(............).))---)).))))......)))).(((((..((.......))..))))).... ( -46.90, z-score = -1.64, R) >droEre2.scaffold_4820 9468762 110 - 10470090 -----ACAGGCUGCCACGUAGGCAUCUACCGCCGCCUGCUUCGCCUCCUCCGUAUACGCUCCAUCGCC---GGUGGCCAUGAAAGCCGCCAAUGCAGCCUUCUCCGGAUCCGUGACUG -----..(((((((...((((((..........))))))..................((......)).---((((((.......))))))...)))))))..((((....)).))... ( -34.10, z-score = -0.57, R) >droYak2.chr3R 27918310 110 + 28832112 -----GCAGGCUGCCACGUAGGCCUCCACCGCCGCCUGUUUGGCCUCCUCCGUGUACGCUCCAUCGCC---GGUGGCCAUGAAAGCCGCCAAUGCAGCCUUUUCCGGAUCCGUGACAG -----..(((((((((((..(((.......)))(((.....)))......))))...((......)).---((((((.......))))))...)))))))..((((....)).))... ( -37.10, z-score = -0.47, R) >droSim1.chr3R 26624984 110 + 27517382 -----ACAGGCCGCCACGUAGGCCUCUACCGCAGCCUUUUUGGCCUCCUCCGUAUACGCUCCAUCGCC---GGUGGCCAUGAAAGCCGCCAAUGCGGCCUUUUCCGGAUCCGUGACAG -----..(((((((...((((....)))).((.((.(((.(((((.((..((............))..---)).))))).))).)).))....)))))))..((((....)).))... ( -36.70, z-score = -0.84, R) >consensus _____ACAGGCCGCCACAUAGGCGGCCACCGCCGCCUGUUUGGCCUCCUCCGUGAUCGCUCCAUCGCC___GGUGGCCACAAAGGCCGCCACUGCAGCCUUUUCCGCAUCCGUGACAG ........((((((...((((((..........)))))).(((((.(.........................).)))))..............))))))................... (-20.39 = -20.76 + 0.37)
| Location | 26,984,207 – 26,984,317 |
|---|---|
| Length | 110 |
| Sequences | 11 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 74.75 |
| Shannon entropy | 0.52003 |
| G+C content | 0.64533 |
| Mean single sequence MFE | -50.96 |
| Consensus MFE | -28.37 |
| Energy contribution | -28.62 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.56 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.923218 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
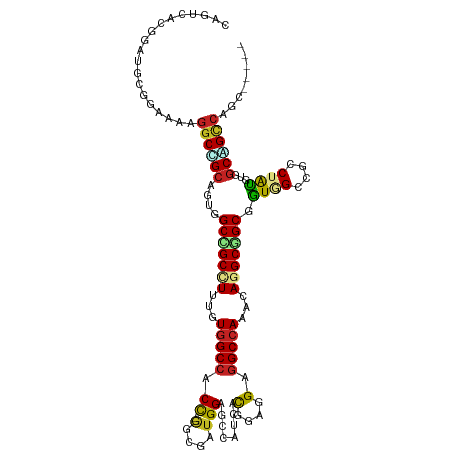
>dm3.chr3R 26984207 110 - 27905053 CUGUCACGGAUCCGGAAAAGGCCGCAUUGGCGGCUUUCAUGGCCACC---GGCGAUGGAGCGUAUACGGAGGAGGCCAAAAAGGCUGCGGUAGAGGCCUACGUGGCGGCCUGU----- ...((.((....))))..(((((((....((((((((..(((((.((---..(((((.....))).))..)).)))))..)))))))).((((....))))...)))))))..----- ( -46.90, z-score = -1.40, R) >droGri2.scaffold_14624 334503 115 + 4233967 CGGUCACCGAUGCGGAGAAGGCGGCUGUCGCUGCCUUUGUGGCCACC---AGCGAUGGCGUCAUCACCGAGGAGGCCAAACAGGCGGCUAUUGCCGCCUAUGUGGCGGCCACUGAGGC .((((.((....(((.((.((((.(((((((((((.....)))....---)))))))))))).)).))).))..((((...(((((((....)))))))...))))))))........ ( -58.10, z-score = -2.62, R) >droMoj3.scaffold_6540 7092033 118 + 34148556 CUGUCACCGAUGCGGAGAAGGCAGCUGUCGCCGCCUUUGUGGCCACCGCAGGCGAAGGCGUCAACAUGGGCGAGGCCAAGCAGGCGGCCAUUGAGGCCUAUGCUGCUGCCCAGGAUGU (((((.(((...))).)).((((((.((....(((((.((((((.((....((...(((.((.........)).)))..)).)).)))))).)))))....)).)))))))))..... ( -50.60, z-score = 0.03, R) >droVir3.scaffold_12855 8842311 115 + 10161210 CAGUAACCGAUGCUGAGAAGGCAGCUGUCGCCGCCUUUAUGGCCACU---GGCGAUGGCGCCAUCACCGACGAGGCCAAACAAGCGGCCGUUGCCGCCUAUGUGGCGGCCAACGAAGC (((((.....))))).((.(((.((((((((((((.....)))....---)))))))))))).))........((((........))))(((((((((.....))))).))))..... ( -53.00, z-score = -2.91, R) >droWil1.scaffold_181108 644503 112 - 4707319 CAGUGACGGAUGCAGAAAAGGCUGCGGUUGCCGCCUUUGUGGCAACU---GCGGAUGGAGCCAUUACCGAGGAGGCCAAACAAGCGGCAGUGGCUGCCUAUCUGGCAGUCAGCAA--- ..........(((.......((((((((((((((....)))))))))---..(..(((..((........))...)))..)..)))))..((((((((.....))))))))))).--- ( -46.60, z-score = -1.46, R) >droPer1.super_7 1502630 110 + 4445127 CAGUCACGGAUGCGGAAAAGGCCGCGGCAGCCGCCUUUGUGGCCGCC---AGCGAUGGGGCAAUCACUGCCGAGGCCAAGCAGGCGGCUGUGGCCGCCUAUGUGGCAGCCAGC----- .......((.(((.(...((((.((.((((((((((...(((((.((---(....)))((((.....))))..)))))...)))))))))).)).))))...).))).))...----- ( -60.70, z-score = -2.52, R) >dp4.chr2 30644536 110 + 30794189 CAGUCACGGAUGCGGAAAAGGCCGCGGCAGCCGCCUUUGUGGCCGCC---AGCGAUGGGGCAAUCACUGCCGAGGCCAAGCAGGCGGCUGUGGCCGCCUAUGUGGCAGCCAGC----- .......((.(((.(...((((.((.((((((((((...(((((.((---(....)))((((.....))))..)))))...)))))))))).)).))))...).))).))...----- ( -60.70, z-score = -2.52, R) >droAna3.scaffold_13340 14366740 112 - 23697760 CUGUAACGGAUGCGGAAAAAGCUGCCGUUGCGGCAUUCGUGGCCACC---AGUGAUGGAGCAAUUACGCCGGAGGCCAAGCAGGCAGCGGUGGCCGCCUGCCCGCCGGACUUUGA--- ..(((((((..((.......))..)))))))(((.....(((((.((---.(((((......)))))...)).))))).((((((.((....)).))))))..))).........--- ( -48.20, z-score = -0.86, R) >droEre2.scaffold_4820 9468762 110 + 10470090 CAGUCACGGAUCCGGAGAAGGCUGCAUUGGCGGCUUUCAUGGCCACC---GGCGAUGGAGCGUAUACGGAGGAGGCGAAGCAGGCGGCGGUAGAUGCCUACGUGGCAGCCUGU----- ..(((.(...((((..(((((((((....)))))))))((.(((...---))).))..........)))).).)))...(((((((.((((((....)))).)).).))))))----- ( -41.20, z-score = -0.06, R) >droYak2.chr3R 27918310 110 - 28832112 CUGUCACGGAUCCGGAAAAGGCUGCAUUGGCGGCUUUCAUGGCCACC---GGCGAUGGAGCGUACACGGAGGAGGCCAAACAGGCGGCGGUGGAGGCCUACGUGGCAGCCUGC----- ...((.((....))))..(((((((..((..(((((((((.(((.((---((((......)))...))).....(((.....)))))).)))))))))..))..)))))))..----- ( -47.70, z-score = -1.41, R) >droSim1.chr3R 26624984 110 - 27517382 CUGUCACGGAUCCGGAAAAGGCCGCAUUGGCGGCUUUCAUGGCCACC---GGCGAUGGAGCGUAUACGGAGGAGGCCAAAAAGGCUGCGGUAGAGGCCUACGUGGCGGCCUGU----- ...((.((....))))..(((((((....((((((((..(((((.((---..(((((.....))).))..)).)))))..)))))))).((((....))))...)))))))..----- ( -46.90, z-score = -1.40, R) >consensus CAGUCACGGAUGCGGAAAAGGCCGCAGUGGCCGCCUUUGUGGCCACC___GGCGAUGGAGCCAUCACGGAGGAGGCCAAACAGGCGGCGGUGGCCGCCUAUGUGGCAGCCAGC_____ ...................((((((....(((((((...(((((.......((......)).....(....).)))))...))))))).((((....))))...))))))........ (-28.37 = -28.62 + 0.25)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:58:59 2011