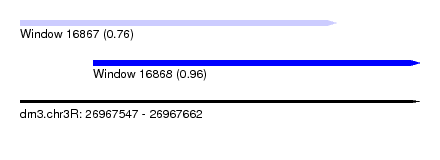
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 26,967,547 – 26,967,662 |
| Length | 115 |
| Max. P | 0.964909 |

| Location | 26,967,547 – 26,967,638 |
|---|---|
| Length | 91 |
| Sequences | 7 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 78.92 |
| Shannon entropy | 0.42015 |
| G+C content | 0.45733 |
| Mean single sequence MFE | -13.74 |
| Consensus MFE | -11.64 |
| Energy contribution | -11.80 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.06 |
| Mean z-score | -0.82 |
| Structure conservation index | 0.85 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.760623 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
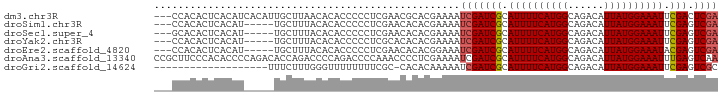
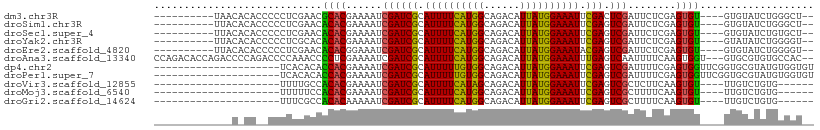
>dm3.chr3R 26967547 91 + 27905053 ---CCACACUCACAUCACAUUGCUUAACACACCCCCUCGAACGCACGAAAAUCGAUCGCAUUUUCAUGGCAGACAUUAUGGAAAUUCGACUCGA ---.................................(((......)))...(((((((.((((((((((......)))))))))).))).)))) ( -14.00, z-score = -1.20, R) >droSim1.chr3R 26608448 86 + 27517382 ---CCACACUCACAU-----UGCUUUACACACCCCCUCGAACACACGAAAAUCGAUCGCAUUUUCAUGGCAGACAUUAUGGAAAUUCGAGUCGA ---............-----................(((......)))...(((((((.((((((((((......)))))))))).))).)))) ( -14.00, z-score = -0.99, R) >droSec1.super_4 5786317 86 + 6179234 ---GCACACUCACAU-----UGCUUUACACACCCCCUCGAACACACGAAAAUCGAUCGCAUUUUCAUGGCAGACAUUAUGGAAAUUCGAGUCGA ---(((.........-----))).............(((......)))...(((((((.((((((((((......)))))))))).))).)))) ( -14.50, z-score = -1.00, R) >droYak2.chr3R 27901250 86 + 28832112 ---CCACACUCACAU-----UGCUUUACACACCCCCUCGCACACACGAAAAUCGAUCGCAUUUUCAUGGCAGACAUUAUGGAAAUUCGAGUCGA ---............-----................(((......)))...(((((((.((((((((((......)))))))))).))).)))) ( -14.00, z-score = -1.03, R) >droEre2.scaffold_4820 9451969 86 - 10470090 ---CCACACUCACAU-----UGCUUUACACACCCCCUCGAACACACGGAAAUCGAUCGCAUUUUCAUGGCAGACAUUAUGGAAAUACGAGUCGA ---............-----...............................(((((((.((((((((((......)))))))))).))).)))) ( -13.10, z-score = -0.41, R) >droAna3.scaffold_13340 14349052 94 + 23697760 CCGCUUCCCACACCCCAGACACCAGACCCCAGACCCCAAACCCCUCGAAAAUCGAUCGCAUUUUCAUGGCAGACAUUAUGGAAAUUUGAGUCAA .................(((........................(((.....)))(((.((((((((((......)))))))))).)))))).. ( -13.10, z-score = -0.54, R) >droGri2.scaffold_14624 312533 74 - 4233967 -------------------UUUCUUUGGGUUUUUUUUCGC-CACACAAAAAUCGAUCGCAUUUUCAUGGCAGACAUUAUGGAAAUUCGAGUCGC -------------------......(((((........))-).)).......((((((.((((((((((......)))))))))).))).))). ( -13.50, z-score = -0.57, R) >consensus ___CCACACUCACAU_____UGCUUUACACACCCCCUCGAACACACGAAAAUCGAUCGCAUUUUCAUGGCAGACAUUAUGGAAAUUCGAGUCGA ...................................................(((((((.((((((((((......)))))))))).))).)))) (-11.64 = -11.80 + 0.16)
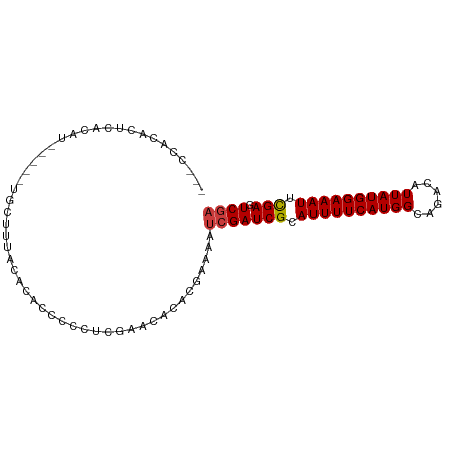
| Location | 26,967,568 – 26,967,662 |
|---|---|
| Length | 94 |
| Sequences | 11 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 74.61 |
| Shannon entropy | 0.47075 |
| G+C content | 0.44409 |
| Mean single sequence MFE | -26.31 |
| Consensus MFE | -14.47 |
| Energy contribution | -13.74 |
| Covariance contribution | -0.73 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.964909 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 26967568 94 + 27905053 ----------UAACACACCCCCUCGAACGCACGAAAAUCGAUCGCAUUUUCAUGGCAGACAUUAUGGAAAUUCGACUCGAUUCUCGAGUGU----GUGUAUCUGGGCU-- ----------.......(((......((((((((.(((((((((.((((((((((......)))))))))).))).)))))).))..))))----))......)))..-- ( -28.80, z-score = -2.54, R) >droSim1.chr3R 26608464 94 + 27517382 ----------UUACACACCCCCUCGAACACACGAAAAUCGAUCGCAUUUUCAUGGCAGACAUUAUGGAAAUUCGAGUCGAUUCUCGAGUGU----GUGUAUCUGGGCU-- ----------.......(((......((((((((.(((((((((.((((((((((......)))))))))).))).)))))).))..))))----))......)))..-- ( -29.20, z-score = -2.55, R) >droSec1.super_4 5786333 94 + 6179234 ----------UUACACACCCCCUCGAACACACGAAAAUCGAUCGCAUUUUCAUGGCAGACAUUAUGGAAAUUCGAGUCGAUUCUCGAGUGU----GUGUAUCUGUGCU-- ----------...((((......((.((((.(((.(((((((((.((((((((((......)))))))))).))).)))))).))).))))----.))....))))..-- ( -28.00, z-score = -2.46, R) >droYak2.chr3R 27901266 94 + 28832112 ----------UUACACACCCCCUCGCACACACGAAAAUCGAUCGCAUUUUCAUGGCAGACAUUAUGGAAAUUCGAGUCGAUUCUCGAGUGU----GUAUAUCUGGGGU-- ----------......(((((...((((((.(((.(((((((((.((((((((((......)))))))))).))).)))))).))).))))----))......)))))-- ( -36.40, z-score = -5.00, R) >droEre2.scaffold_4820 9451985 94 - 10470090 ----------UUACACACCCCCUCGAACACACGGAAAUCGAUCGCAUUUUCAUGGCAGACAUUAUGGAAAUACGAGUCGAUUCUCGAGUGU----GUGUAUCUGGGGU-- ----------......(((((.....((((((.(((((((((((.((((((((((......)))))))))).))).)))))).))..))))----))......)))))-- ( -34.00, z-score = -3.39, R) >droAna3.scaffold_13340 14349066 105 + 23697760 CCAGACACCAGACCCCAGACCCCAAACCCCUCGAAAAUCGAUCGCAUUUUCAUGGCAGACAUUAUGGAAAUUUGAGUCAAUUUUCAAGUGGU---GUGCGUGUGCCAC-- ....((((((.(((.(.((...........))((((((.(((((.((((((((((......)))))))))).))).)).))))))..).)))---.)).)))).....-- ( -23.30, z-score = -0.66, R) >dp4.chr2 30620664 89 - 30794189 ---------------------UCACACACCACGAAAAUCGAUCGCAUUUUUGUGGCAGACAUUAUGGAAAUUCGAGUCGAUUUUCGAGUGGUUCGGUGCGUAUGUGGUGU ---------------------....(((((((((((((((((((.(((((..(((......)))..))))).))).)))))))))..((........))....))))))) ( -29.80, z-score = -2.26, R) >droPer1.super_7 1486170 89 - 4445127 ---------------------UCACACACCACGAAAAUCGAUCGCAUUUUUGUGGCAGACAUUAUGGAAAUUCGAGUCGAUUUUCGAGUGGUUCGGUGCGUAUGUGGUGU ---------------------....(((((((((((((((((((.(((((..(((......)))..))))).))).)))))))))..((........))....))))))) ( -29.80, z-score = -2.26, R) >droVir3.scaffold_12855 8820747 79 - 10161210 ---------------------UUUUGCCACACGAAAAUCGAUCGCAUUUUCAUAGCAGACAUUAUGGAAAUUCGAGUCGCUCUUCAAGUGU----UUGUCUGUG------ ---------------------......(((..(((...((((((.((((((((((......)))))))))).))).)))...)))..))).----.........------ ( -15.50, z-score = -0.14, R) >droMoj3.scaffold_6540 7069801 79 - 34148556 ---------------------UUUUUCCACACGAAAAUCGAUCGCAUUUUCAUGGCAGACAUUAUGGAAAUUCGAGUCGCUUUUCAAGUGU----UUGUCUGUG------ ---------------------......((((.(((((((....).))))))..((((((((((...((((..((...))..)))).)))))----)))))))))------ ( -18.70, z-score = -1.37, R) >droGri2.scaffold_14624 312548 79 - 4233967 ---------------------UUUCGCCACACAAAAAUCGAUCGCAUUUUCAUGGCAGACAUUAUGGAAAUUCGAGUCGCUUUUCAAGUGU----UUGUCUGUG------ ---------------------........((((......((........))..((((((((((...((((..((...))..)))).)))))----)))))))))------ ( -15.90, z-score = -0.35, R) >consensus _____________C_CA__CCCUCGACCACACGAAAAUCGAUCGCAUUUUCAUGGCAGACAUUAUGGAAAUUCGAGUCGAUUUUCGAGUGU____GUGUAUCUGGGGU__ ............................((((......((((((.((((((((((......)))))))))).))).)))........))))................... (-14.47 = -13.74 + -0.73)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:58:56 2011